|
probeID |
AGICode |
Annotation |
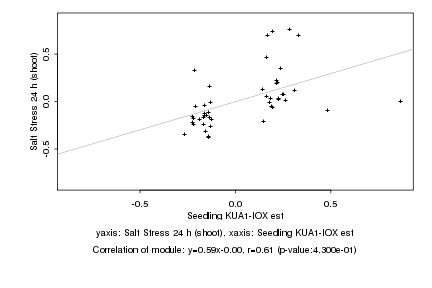
Log2 signal ratio
Seedling KUA1-IOX est |
Log2 signal ratio
Salt Stress 24 h (shoot) |
| 1 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.863 |
0.006 |
| 2 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.482 |
-0.092 |
| 3 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.328 |
0.702 |
| 4 |
264314_at |
AT1G70420
|
unknown |
0.309 |
0.117 |
| 5 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.281 |
0.758 |
| 6 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.258 |
0.021 |
| 7 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.249 |
0.074 |
| 8 |
264951_at |
AT1G76970
|
[Target of Myb protein 1] |
0.244 |
0.076 |
| 9 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.232 |
0.348 |
| 10 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.221 |
0.024 |
| 11 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.221 |
0.041 |
| 12 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.216 |
0.202 |
| 13 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.210 |
0.228 |
| 14 |
255462_at |
AT4G02940
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.210 |
0.199 |
| 15 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.192 |
0.738 |
| 16 |
265539_at |
AT2G15830
|
unknown |
0.190 |
-0.054 |
| 17 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
0.188 |
-0.044 |
| 18 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.179 |
0.040 |
| 19 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.176 |
-0.000 |
| 20 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.163 |
0.704 |
| 21 |
246631_at |
AT1G50740
|
[Transmembrane proteins 14C] |
0.161 |
0.061 |
| 22 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.161 |
0.465 |
| 23 |
261567_at |
AT1G33055
|
unknown |
0.145 |
-0.205 |
| 24 |
261545_at |
AT1G63530
|
unknown |
0.141 |
0.133 |
| 25 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.269 |
-0.343 |
| 26 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.227 |
-0.213 |
| 27 |
248329_at |
AT5G52780
|
unknown |
-0.226 |
-0.150 |
| 28 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.224 |
-0.169 |
| 29 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.220 |
-0.241 |
| 30 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.215 |
0.330 |
| 31 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.213 |
-0.043 |
| 32 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.193 |
-0.183 |
| 33 |
254612_at |
AT4G19100
|
PAM68, photosynthesis affected mutant 68 |
-0.172 |
-0.163 |
| 34 |
248920_at |
AT5G45930
|
CHL I2, CHLI-2, CHLI2, magnesium chelatase i2 |
-0.171 |
-0.239 |
| 35 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.165 |
-0.137 |
| 36 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.165 |
-0.117 |
| 37 |
249482_at |
AT5G38980
|
unknown |
-0.163 |
-0.039 |
| 38 |
266707_at |
AT2G03310
|
unknown |
-0.161 |
-0.306 |
| 39 |
254911_at |
AT4G11100
|
unknown |
-0.152 |
-0.137 |
| 40 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
-0.145 |
-0.367 |
| 41 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.142 |
-0.371 |
| 42 |
255436_at |
AT4G03150
|
unknown |
-0.142 |
-0.114 |
| 43 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
-0.140 |
-0.159 |
| 44 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.139 |
0.159 |
| 45 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.135 |
-0.007 |
| 46 |
266329_at |
AT2G01590
|
CRR3, CHLORORESPIRATORY REDUCTION 3 |
-0.135 |
-0.261 |
| 47 |
262648_at |
AT1G14030
|
LSMT-L, lysine methyltransferase (LSMT)-like |
-0.130 |
-0.180 |