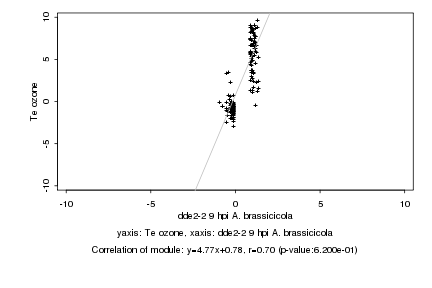
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
dde2-2 9 hpi A. brassicicola |
Log2 signal ratio
Te ozone |
| 1 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.320 |
2.417 |
| 2 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
1.310 |
1.575 |
| 3 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
1.307 |
5.260 |
| 4 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
1.284 |
9.729 |
| 5 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.272 |
1.246 |
| 6 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
1.270 |
8.860 |
| 7 |
253044_at |
AT4G37290
|
unknown |
1.233 |
5.891 |
| 8 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
1.210 |
6.732 |
| 9 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.190 |
2.310 |
| 10 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.185 |
4.628 |
| 11 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
1.180 |
8.685 |
| 12 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
1.170 |
7.758 |
| 13 |
253796_at |
AT4G28460
|
unknown |
1.169 |
6.687 |
| 14 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.164 |
6.327 |
| 15 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.162 |
5.944 |
| 16 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
1.157 |
7.802 |
| 17 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
1.134 |
-0.428 |
| 18 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
1.131 |
7.113 |
| 19 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.121 |
9.091 |
| 20 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
1.121 |
7.917 |
| 21 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.104 |
5.551 |
| 22 |
264580_at |
AT1G05340
|
unknown |
1.103 |
7.166 |
| 23 |
246370_at |
AT1G51920
|
unknown |
1.102 |
7.594 |
| 24 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
1.101 |
9.090 |
| 25 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
1.092 |
6.405 |
| 26 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
1.079 |
6.570 |
| 27 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
1.061 |
7.002 |
| 28 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
1.044 |
3.484 |
| 29 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.039 |
3.349 |
| 30 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
1.038 |
8.270 |
| 31 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.036 |
2.476 |
| 32 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
1.022 |
8.157 |
| 33 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.022 |
1.776 |
| 34 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
1.004 |
8.491 |
| 35 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.988 |
4.963 |
| 36 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.987 |
5.372 |
| 37 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.984 |
8.650 |
| 38 |
245082_at |
AT2G23270
|
unknown |
0.980 |
6.731 |
| 39 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.980 |
1.348 |
| 40 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.973 |
2.843 |
| 41 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.972 |
8.548 |
| 42 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.968 |
3.793 |
| 43 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.967 |
8.740 |
| 44 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.963 |
1.163 |
| 45 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.955 |
5.770 |
| 46 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.951 |
4.940 |
| 47 |
258203_at |
AT3G13950
|
unknown |
0.946 |
6.661 |
| 48 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.946 |
8.790 |
| 49 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.945 |
3.047 |
| 50 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.933 |
5.459 |
| 51 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.931 |
6.705 |
| 52 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.928 |
3.752 |
| 53 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.927 |
4.836 |
| 54 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.927 |
6.714 |
| 55 |
256442_at |
AT3G10930
|
unknown |
0.923 |
5.177 |
| 56 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.919 |
4.323 |
| 57 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.919 |
3.457 |
| 58 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.917 |
7.358 |
| 59 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.911 |
7.296 |
| 60 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.910 |
6.660 |
| 61 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.908 |
3.786 |
| 62 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.894 |
8.419 |
| 63 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.887 |
7.512 |
| 64 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.884 |
9.031 |
| 65 |
254249_at |
AT4G23280
|
CRK20, cysteine-rich RLK (RECEPTOR-like protein kinase) 20 |
0.883 |
5.856 |
| 66 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.873 |
8.874 |
| 67 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.869 |
6.648 |
| 68 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.869 |
2.582 |
| 69 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.869 |
7.446 |
| 70 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.863 |
1.369 |
| 71 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.862 |
4.445 |
| 72 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.862 |
7.480 |
| 73 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.859 |
6.025 |
| 74 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.858 |
5.838 |
| 75 |
245755_at |
AT1G35210
|
unknown |
0.855 |
5.757 |
| 76 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.852 |
8.282 |
| 77 |
260754_at |
AT1G49000
|
unknown |
0.850 |
5.674 |
| 78 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.848 |
4.658 |
| 79 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.840 |
7.475 |
| 80 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.993 |
-0.043 |
| 81 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.788 |
-0.530 |
| 82 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.787 |
-0.544 |
| 83 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.568 |
-1.048 |
| 84 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.554 |
-0.041 |
| 85 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.552 |
3.405 |
| 86 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.543 |
-2.399 |
| 87 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.538 |
-0.734 |
| 88 |
256603_at |
AT3G28270
|
unknown |
-0.481 |
-1.603 |
| 89 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.478 |
-1.110 |
| 90 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.458 |
3.489 |
| 91 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.434 |
-1.092 |
| 92 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.433 |
0.769 |
| 93 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.401 |
0.326 |
| 94 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.347 |
-1.952 |
| 95 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.346 |
2.276 |
| 96 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
-0.344 |
-0.890 |
| 97 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.342 |
-0.214 |
| 98 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-0.324 |
-0.808 |
| 99 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
-0.317 |
-1.185 |
| 100 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.301 |
0.691 |
| 101 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.297 |
-0.675 |
| 102 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
-0.295 |
-0.463 |
| 103 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.289 |
0.118 |
| 104 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.287 |
-1.648 |
| 105 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.285 |
-1.997 |
| 106 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.277 |
-0.355 |
| 107 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.270 |
-0.637 |
| 108 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.266 |
-1.971 |
| 109 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.264 |
-0.673 |
| 110 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.263 |
-0.675 |
| 111 |
251852_at |
AT3G54750
|
unknown |
-0.255 |
-0.250 |
| 112 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
-0.246 |
-0.258 |
| 113 |
257024_at |
AT3G19100
|
[Protein kinase superfamily protein] |
-0.240 |
-0.265 |
| 114 |
258326_at |
AT3G22760
|
SOL1 |
-0.232 |
-0.989 |
| 115 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.224 |
-1.913 |
| 116 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.205 |
-0.307 |
| 117 |
253351_at |
AT4G33700
|
unknown |
-0.202 |
-0.947 |
| 118 |
254764_at |
AT4G13250
|
NYC1, NON-YELLOW COLORING 1 |
-0.201 |
-1.132 |
| 119 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.193 |
-1.252 |
| 120 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
-0.192 |
-1.238 |
| 121 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.190 |
-0.798 |
| 122 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
-0.183 |
-0.473 |
| 123 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.182 |
-0.413 |
| 124 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.177 |
-0.946 |
| 125 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.176 |
-1.222 |
| 126 |
249058_at |
AT5G44510
|
TAO1, target of AVRB operation1 |
-0.176 |
-1.266 |
| 127 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
-0.171 |
-1.790 |
| 128 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.166 |
-2.867 |
| 129 |
258587_at |
AT3G04310
|
unknown |
-0.165 |
-0.643 |
| 130 |
258051_at |
AT3G16260
|
TRZ4, tRNAse Z4 |
-0.163 |
-1.307 |
| 131 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.163 |
-0.518 |
| 132 |
258089_at |
AT3G14740
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.161 |
-0.355 |
| 133 |
252194_at |
AT3G50110
|
ATPEN3, Arabidopsis thaliana phosphatase and TENsin homolog deleted on chromosome ten 3, PEN3, PTEN 3, PTEN2B, phosphatase and TENsin homolog deleted on chromosome ten 2B |
-0.161 |
-0.060 |
| 134 |
264604_at |
AT1G04650
|
unknown |
-0.160 |
-1.097 |
| 135 |
248155_at |
AT5G54390
|
AHL, HAL2-like, ATAHL, HL, HAL2-like |
-0.158 |
0.786 |
| 136 |
250079_at |
AT5G16650
|
[Chaperone DnaJ-domain superfamily protein] |
-0.158 |
-0.755 |
| 137 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.155 |
-1.109 |
| 138 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.155 |
-0.580 |
| 139 |
245598_at |
AT4G14200
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.150 |
-0.299 |
| 140 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
-0.150 |
-0.913 |
| 141 |
262854_at |
AT1G20870
|
[HSP20-like chaperones superfamily protein] |
-0.148 |
-1.618 |
| 142 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.148 |
-0.745 |
| 143 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.147 |
-0.871 |
| 144 |
260260_at |
AT1G68540
|
CCRL6, cinnamoyl coA reductase-like 6, TKPR2, tetraketide alpha-pyrone reductase 2 |
-0.145 |
-1.063 |
| 145 |
250074_at |
AT5G17310
|
AtUGP2, UDP-GLUCOSE PYROPHOSPHORYLASE 2, UGP2, UDP-glucose pyrophosphorylase 2 |
-0.145 |
-0.515 |
| 146 |
257265_at |
AT3G14980
|
IDM1, increased DNA methylation 1, ROS4, REPRESSOR OF SILENCING 4 |
-0.142 |
-1.101 |
| 147 |
247716_at |
AT5G59350
|
unknown |
-0.141 |
-2.291 |
| 148 |
253022_at |
AT4G38060
|
CCI2, Clavata complex interactor 2 |
-0.139 |
-0.333 |
| 149 |
252785_at |
AT3G42660
|
[transducin family protein / WD-40 repeat family protein] |
-0.136 |
-0.484 |
| 150 |
254026_at |
AT4G25800
|
[Calmodulin-binding protein] |
-0.135 |
-0.659 |
| 151 |
262292_at |
AT1G27595
|
unknown |
-0.135 |
-0.172 |
| 152 |
261871_at |
AT1G11440
|
unknown |
-0.135 |
-1.857 |
| 153 |
258149_at |
AT3G18110
|
EMB1270, embryo defective 1270 |
-0.134 |
-1.390 |
| 154 |
257855_at |
AT3G13040
|
[myb-like HTH transcriptional regulator family protein] |
-0.134 |
-1.195 |
| 155 |
247466_at |
AT5G62090
|
SLK2, SEUSS-like 2 |
-0.132 |
-0.990 |
| 156 |
246948_at |
AT5G25130
|
CYP71B12, cytochrome P450, family 71, subfamily B, polypeptide 12 |
-0.130 |
-2.035 |
| 157 |
265570_at |
AT2G28310
|
unknown |
-0.129 |
-1.436 |