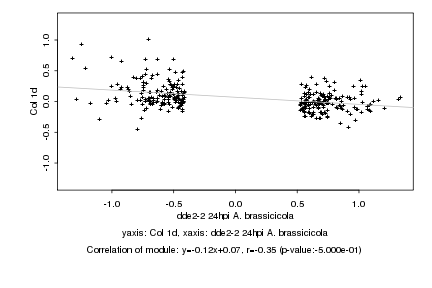
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
dde2-2 24hpi A. brassicicola |
Log2 signal ratio
Col 1d |
| 1 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
1.333 |
0.069 |
| 2 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
1.317 |
0.037 |
| 3 |
249454_at |
AT5G39520
|
unknown |
1.206 |
-0.106 |
| 4 |
251755_at |
AT3G55790
|
unknown |
1.156 |
0.026 |
| 5 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
1.115 |
0.015 |
| 6 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
1.092 |
-0.047 |
| 7 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
1.089 |
-0.162 |
| 8 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
1.085 |
-0.063 |
| 9 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
1.080 |
-0.134 |
| 10 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
1.066 |
-0.125 |
| 11 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
1.065 |
-0.074 |
| 12 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
1.062 |
-0.079 |
| 13 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
1.049 |
0.253 |
| 14 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.026 |
-0.009 |
| 15 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
1.023 |
0.247 |
| 16 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
1.018 |
0.126 |
| 17 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
1.017 |
0.174 |
| 18 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
1.013 |
-0.175 |
| 19 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
1.007 |
0.352 |
| 20 |
256603_at |
AT3G28270
|
unknown |
0.985 |
-0.116 |
| 21 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.972 |
-0.124 |
| 22 |
265892_at |
AT2G15020
|
unknown |
0.969 |
-0.293 |
| 23 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.962 |
0.059 |
| 24 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.960 |
-0.089 |
| 25 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.952 |
0.248 |
| 26 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.929 |
0.034 |
| 27 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
0.927 |
-0.210 |
| 28 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.918 |
0.079 |
| 29 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.909 |
-0.417 |
| 30 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.907 |
0.067 |
| 31 |
257057_at |
AT3G15310
|
unknown |
0.903 |
-0.153 |
| 32 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.874 |
-0.060 |
| 33 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
0.865 |
-0.118 |
| 34 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.863 |
0.091 |
| 35 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.857 |
-0.105 |
| 36 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.855 |
0.010 |
| 37 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.850 |
-0.355 |
| 38 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.849 |
-0.069 |
| 39 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.836 |
0.064 |
| 40 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.835 |
-0.056 |
| 41 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.824 |
-0.110 |
| 42 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.817 |
-0.087 |
| 43 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.816 |
0.063 |
| 44 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.813 |
0.047 |
| 45 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.808 |
0.061 |
| 46 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.799 |
0.181 |
| 47 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
0.795 |
0.317 |
| 48 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.779 |
-0.038 |
| 49 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.774 |
0.108 |
| 50 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.768 |
0.064 |
| 51 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.761 |
0.074 |
| 52 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.758 |
0.132 |
| 53 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.757 |
0.255 |
| 54 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.748 |
-0.032 |
| 55 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.747 |
0.034 |
| 56 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.745 |
-0.137 |
| 57 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.745 |
-0.025 |
| 58 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.744 |
0.086 |
| 59 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.742 |
-0.250 |
| 60 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.742 |
-0.237 |
| 61 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.738 |
-0.012 |
| 62 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.732 |
0.326 |
| 63 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.731 |
-0.023 |
| 64 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.728 |
0.028 |
| 65 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.722 |
-0.184 |
| 66 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.721 |
0.044 |
| 67 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.718 |
0.381 |
| 68 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.717 |
0.074 |
| 69 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
0.714 |
-0.165 |
| 70 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.713 |
0.089 |
| 71 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.711 |
0.004 |
| 72 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.711 |
-0.060 |
| 73 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.710 |
0.116 |
| 74 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.709 |
-0.092 |
| 75 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.705 |
0.014 |
| 76 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
0.699 |
0.096 |
| 77 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.697 |
-0.044 |
| 78 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.697 |
-0.205 |
| 79 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.695 |
-0.171 |
| 80 |
247055_at |
AT5G66740
|
unknown |
0.692 |
0.106 |
| 81 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.684 |
-0.029 |
| 82 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
0.684 |
-0.270 |
| 83 |
251612_at |
AT3G57950
|
unknown |
0.683 |
-0.168 |
| 84 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.683 |
-0.010 |
| 85 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.682 |
0.135 |
| 86 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
0.680 |
-0.004 |
| 87 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.677 |
0.048 |
| 88 |
254145_at |
AT4G24700
|
unknown |
0.674 |
-0.268 |
| 89 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.673 |
-0.033 |
| 90 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.669 |
-0.081 |
| 91 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.666 |
0.050 |
| 92 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.666 |
-0.108 |
| 93 |
250926_at |
AT5G03555
|
AtNCS1, NCS1, nucleobase cation symporter 1 |
0.665 |
-0.001 |
| 94 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.662 |
0.030 |
| 95 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.661 |
-0.000 |
| 96 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.654 |
-0.265 |
| 97 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.654 |
-0.061 |
| 98 |
246125_at |
AT5G19875
|
unknown |
0.654 |
0.149 |
| 99 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.653 |
0.280 |
| 100 |
259856_at |
AT1G68440
|
unknown |
0.642 |
-0.047 |
| 101 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.637 |
-0.011 |
| 102 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.633 |
-0.001 |
| 103 |
255763_at |
AT1G16730
|
unknown |
0.630 |
-0.172 |
| 104 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.630 |
0.014 |
| 105 |
251727_at |
AT3G56290
|
unknown |
0.630 |
-0.084 |
| 106 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.624 |
-0.198 |
| 107 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.624 |
0.117 |
| 108 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.620 |
-0.010 |
| 109 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.619 |
-0.207 |
| 110 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.614 |
-0.231 |
| 111 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.608 |
0.401 |
| 112 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.605 |
0.116 |
| 113 |
260875_at |
AT1G21410
|
SKP2A |
0.605 |
-0.034 |
| 114 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
0.604 |
-0.035 |
| 115 |
265387_at |
AT2G20670
|
unknown |
0.599 |
-0.100 |
| 116 |
254693_at |
AT4G17880
|
MYC4 |
0.596 |
-0.194 |
| 117 |
256096_at |
AT1G13650
|
unknown |
0.593 |
0.074 |
| 118 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
0.593 |
-0.173 |
| 119 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
0.593 |
0.206 |
| 120 |
247882_at |
AT5G57785
|
unknown |
0.591 |
0.067 |
| 121 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.589 |
0.147 |
| 122 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.586 |
-0.014 |
| 123 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
0.586 |
-0.174 |
| 124 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.584 |
-0.063 |
| 125 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
0.575 |
-0.014 |
| 126 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.572 |
0.038 |
| 127 |
256861_at |
AT3G23920
|
AtBAM1, BAM1, beta-amylase 1, BMY7, BETA-AMYLASE 7, TR-BAMY |
0.571 |
0.119 |
| 128 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.570 |
0.264 |
| 129 |
267178_at |
AT2G37750
|
unknown |
0.568 |
0.017 |
| 130 |
257186_at |
AT3G13130
|
unknown |
0.566 |
-0.037 |
| 131 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.566 |
-0.043 |
| 132 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.566 |
0.230 |
| 133 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.565 |
-0.046 |
| 134 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.565 |
-0.236 |
| 135 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.563 |
-0.099 |
| 136 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.562 |
0.028 |
| 137 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.560 |
-0.072 |
| 138 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.559 |
-0.006 |
| 139 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
0.559 |
-0.167 |
| 140 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.558 |
-0.086 |
| 141 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.558 |
-0.017 |
| 142 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.557 |
0.097 |
| 143 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.556 |
0.142 |
| 144 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.555 |
-0.084 |
| 145 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.554 |
-0.158 |
| 146 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
0.552 |
-0.242 |
| 147 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
0.550 |
-0.113 |
| 148 |
245056_at |
AT2G26480
|
UGT76D1, UDP-glucosyl transferase 76D1 |
0.542 |
-0.143 |
| 149 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.539 |
-0.123 |
| 150 |
258362_at |
AT3G14280
|
unknown |
0.536 |
-0.100 |
| 151 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.534 |
-0.017 |
| 152 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.534 |
-0.009 |
| 153 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.533 |
-0.129 |
| 154 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.530 |
0.290 |
| 155 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.528 |
0.053 |
| 156 |
265913_at |
AT2G25625
|
unknown |
0.527 |
-0.003 |
| 157 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.524 |
-0.131 |
| 158 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
0.523 |
-0.056 |
| 159 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.522 |
-0.036 |
| 160 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-1.321 |
0.704 |
| 161 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-1.288 |
0.045 |
| 162 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.253 |
0.943 |
| 163 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-1.221 |
0.548 |
| 164 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-1.175 |
-0.025 |
| 165 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-1.103 |
-0.289 |
| 166 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-1.044 |
-0.025 |
| 167 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-1.033 |
0.024 |
| 168 |
253322_at |
AT4G33980
|
unknown |
-1.007 |
0.724 |
| 169 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
-1.004 |
0.249 |
| 170 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.978 |
0.062 |
| 171 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.970 |
0.010 |
| 172 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.962 |
0.281 |
| 173 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
-0.932 |
0.199 |
| 174 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.927 |
0.664 |
| 175 |
267036_at |
AT2G38465
|
unknown |
-0.923 |
0.241 |
| 176 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.875 |
0.237 |
| 177 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.873 |
0.196 |
| 178 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.864 |
0.167 |
| 179 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.855 |
0.083 |
| 180 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.842 |
-0.037 |
| 181 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.832 |
0.391 |
| 182 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.809 |
0.380 |
| 183 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.799 |
0.024 |
| 184 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.796 |
-0.441 |
| 185 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.775 |
0.376 |
| 186 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
-0.774 |
0.026 |
| 187 |
246929_at |
AT5G25210
|
unknown |
-0.764 |
-0.266 |
| 188 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.764 |
0.136 |
| 189 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.757 |
-0.043 |
| 190 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
-0.756 |
0.422 |
| 191 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.754 |
0.076 |
| 192 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
-0.750 |
0.190 |
| 193 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.745 |
0.311 |
| 194 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.745 |
0.313 |
| 195 |
260688_at |
AT1G17665
|
unknown |
-0.745 |
0.274 |
| 196 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.745 |
0.232 |
| 197 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.739 |
-0.141 |
| 198 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
-0.735 |
0.043 |
| 199 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.734 |
0.689 |
| 200 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.734 |
0.005 |
| 201 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.733 |
0.453 |
| 202 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
-0.730 |
0.004 |
| 203 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.726 |
0.527 |
| 204 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.724 |
-0.105 |
| 205 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.721 |
0.307 |
| 206 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.711 |
0.022 |
| 207 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.707 |
0.058 |
| 208 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.704 |
1.009 |
| 209 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.701 |
0.150 |
| 210 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.698 |
0.050 |
| 211 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.698 |
0.016 |
| 212 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.695 |
0.015 |
| 213 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
-0.693 |
-0.029 |
| 214 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.689 |
0.153 |
| 215 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.688 |
-0.045 |
| 216 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.685 |
0.376 |
| 217 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.684 |
0.079 |
| 218 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.678 |
0.434 |
| 219 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.676 |
-0.039 |
| 220 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
-0.671 |
0.064 |
| 221 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
-0.671 |
0.030 |
| 222 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.666 |
-0.005 |
| 223 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.647 |
0.157 |
| 224 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
-0.644 |
-0.013 |
| 225 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.639 |
0.006 |
| 226 |
261318_at |
AT1G53035
|
unknown |
-0.638 |
0.444 |
| 227 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.636 |
0.689 |
| 228 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.633 |
0.187 |
| 229 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.631 |
0.061 |
| 230 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
-0.621 |
0.111 |
| 231 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.610 |
-0.122 |
| 232 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.600 |
0.165 |
| 233 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.600 |
-0.054 |
| 234 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.599 |
-0.058 |
| 235 |
257918_at |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
-0.598 |
0.092 |
| 236 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
-0.596 |
-0.005 |
| 237 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.593 |
-0.032 |
| 238 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.589 |
0.248 |
| 239 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.582 |
-0.026 |
| 240 |
245188_at |
AT1G67660
|
[Restriction endonuclease, type II-like superfamily protein] |
-0.581 |
-0.047 |
| 241 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.581 |
0.114 |
| 242 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.578 |
0.047 |
| 243 |
254645_at |
AT4G18520
|
SEL1, SEEDLING LETHAL 1 |
-0.572 |
0.086 |
| 244 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.559 |
0.201 |
| 245 |
266670_at |
AT2G29740
|
UGT71C2, UDP-glucosyl transferase 71C2 |
-0.553 |
0.136 |
| 246 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.547 |
-0.159 |
| 247 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.547 |
-0.012 |
| 248 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.547 |
0.365 |
| 249 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-0.546 |
0.045 |
| 250 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.545 |
0.069 |
| 251 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
-0.540 |
0.336 |
| 252 |
250127_at |
AT5G16380
|
unknown |
-0.539 |
0.086 |
| 253 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.535 |
0.102 |
| 254 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
-0.535 |
0.524 |
| 255 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.534 |
-0.035 |
| 256 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.533 |
0.158 |
| 257 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
-0.532 |
0.084 |
| 258 |
260411_at |
AT1G69890
|
unknown |
-0.530 |
0.292 |
| 259 |
262452_at |
AT1G11210
|
unknown |
-0.526 |
0.279 |
| 260 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.522 |
0.265 |
| 261 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.515 |
-0.095 |
| 262 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.515 |
0.232 |
| 263 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.515 |
0.200 |
| 264 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.509 |
0.128 |
| 265 |
263032_at |
AT1G23850
|
unknown |
-0.503 |
0.258 |
| 266 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.503 |
0.239 |
| 267 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.502 |
0.690 |
| 268 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.499 |
0.049 |
| 269 |
264987_at |
AT1G27030
|
unknown |
-0.498 |
0.278 |
| 270 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.495 |
0.063 |
| 271 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
-0.495 |
0.072 |
| 272 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
-0.493 |
-0.009 |
| 273 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
-0.491 |
0.021 |
| 274 |
258935_at |
AT3G10120
|
unknown |
-0.491 |
0.478 |
| 275 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.487 |
0.055 |
| 276 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
-0.485 |
0.240 |
| 277 |
245600_at |
AT4G14230
|
unknown |
-0.482 |
0.019 |
| 278 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
-0.481 |
0.081 |
| 279 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.475 |
0.163 |
| 280 |
266229_at |
AT2G28840
|
XBAT31, XB3 ortholog 1 in Arabidopsis thaliana |
-0.473 |
0.091 |
| 281 |
255942_at |
AT1G22360
|
AtUGT85A2, UDP-glucosyl transferase 85A2, UGT85A2, UDP-glucosyl transferase 85A2 |
-0.472 |
0.279 |
| 282 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.467 |
0.136 |
| 283 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.467 |
-0.015 |
| 284 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.465 |
0.343 |
| 285 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.464 |
-0.089 |
| 286 |
259001_at |
AT3G01960
|
unknown |
-0.463 |
0.107 |
| 287 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.463 |
0.050 |
| 288 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.463 |
-0.105 |
| 289 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.462 |
-0.107 |
| 290 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.459 |
0.042 |
| 291 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-0.457 |
0.227 |
| 292 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.453 |
-0.068 |
| 293 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.452 |
-0.017 |
| 294 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.447 |
-0.031 |
| 295 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.444 |
0.177 |
| 296 |
258485_at |
AT3G02630
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
-0.442 |
-0.064 |
| 297 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
-0.440 |
0.020 |
| 298 |
263912_at |
AT2G36390
|
BE3, BRANCHING ENZYME 3, SBE2.1, starch branching enzyme 2.1 |
-0.438 |
-0.002 |
| 299 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.437 |
-0.028 |
| 300 |
249990_at |
AT5G18540
|
unknown |
-0.436 |
0.111 |
| 301 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.435 |
0.478 |
| 302 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.435 |
-0.020 |
| 303 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
-0.434 |
0.043 |
| 304 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
-0.434 |
-0.017 |
| 305 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.433 |
0.288 |
| 306 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.432 |
0.397 |
| 307 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.432 |
-0.121 |
| 308 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.430 |
-0.159 |
| 309 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
-0.427 |
0.494 |
| 310 |
261662_at |
AT1G18350
|
ATMKK7, MAP kinase kinase 7, BUD1, BUSHY AND DWARF 1, MKK7, MAP kinase kinase 7, MKK7, MAP KINASE KINASE7 |
-0.425 |
-0.012 |
| 311 |
259118_at |
AT3G01310
|
[Phosphoglycerate mutase-like family protein] |
-0.423 |
0.037 |
| 312 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.422 |
0.004 |
| 313 |
260028_at |
AT1G29980
|
unknown |
-0.422 |
0.139 |
| 314 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.421 |
0.011 |
| 315 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
-0.417 |
0.066 |
| 316 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
-0.416 |
0.156 |
| 317 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.414 |
0.171 |