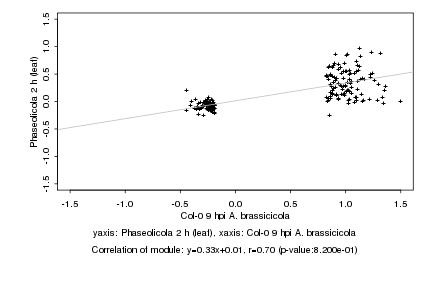
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 9 hpi A. brassicicola |
Log2 signal ratio
Phaseolicola 2 h (leaf) |
| 1 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
1.494 |
0.009 |
| 2 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
1.355 |
0.288 |
| 3 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.350 |
0.217 |
| 4 |
253044_at |
AT4G37290
|
unknown |
1.337 |
-0.018 |
| 5 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
1.327 |
0.076 |
| 6 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
1.310 |
0.884 |
| 7 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
1.297 |
0.312 |
| 8 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.283 |
0.031 |
| 9 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.254 |
0.394 |
| 10 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
1.237 |
0.515 |
| 11 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
1.234 |
0.896 |
| 12 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
1.225 |
0.498 |
| 13 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.225 |
0.441 |
| 14 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
1.212 |
0.043 |
| 15 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.169 |
0.418 |
| 16 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
1.153 |
0.022 |
| 17 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
1.148 |
0.015 |
| 18 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
1.147 |
0.420 |
| 19 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
1.143 |
0.134 |
| 20 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
1.133 |
0.412 |
| 21 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
1.132 |
0.836 |
| 22 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.124 |
0.979 |
| 23 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
1.119 |
0.656 |
| 24 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
1.112 |
0.575 |
| 25 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.105 |
0.667 |
| 26 |
253796_at |
AT4G28460
|
unknown |
1.102 |
0.235 |
| 27 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
1.101 |
0.376 |
| 28 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.097 |
0.550 |
| 29 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
1.094 |
0.745 |
| 30 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.093 |
0.034 |
| 31 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
1.090 |
0.085 |
| 32 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.084 |
0.087 |
| 33 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.080 |
0.516 |
| 34 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
1.079 |
-0.013 |
| 35 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.051 |
0.256 |
| 36 |
264580_at |
AT1G05340
|
unknown |
1.051 |
0.155 |
| 37 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
1.049 |
0.377 |
| 38 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
1.043 |
0.523 |
| 39 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
1.036 |
0.343 |
| 40 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
1.029 |
0.580 |
| 41 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.028 |
0.506 |
| 42 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
1.027 |
0.413 |
| 43 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.027 |
0.198 |
| 44 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
1.026 |
0.037 |
| 45 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.020 |
0.206 |
| 46 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
1.017 |
0.022 |
| 47 |
245082_at |
AT2G23270
|
unknown |
1.017 |
-0.028 |
| 48 |
254408_at |
AT4G21390
|
B120 |
1.014 |
0.357 |
| 49 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
1.014 |
0.865 |
| 50 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
1.013 |
0.209 |
| 51 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
1.005 |
0.850 |
| 52 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
1.002 |
0.561 |
| 53 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
1.001 |
0.189 |
| 54 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.998 |
0.279 |
| 55 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.995 |
0.296 |
| 56 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.993 |
0.565 |
| 57 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.988 |
0.120 |
| 58 |
246370_at |
AT1G51920
|
unknown |
0.987 |
0.700 |
| 59 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.982 |
0.155 |
| 60 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.979 |
0.556 |
| 61 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.978 |
0.134 |
| 62 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.975 |
0.288 |
| 63 |
258203_at |
AT3G13950
|
unknown |
0.972 |
0.426 |
| 64 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.966 |
0.223 |
| 65 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.960 |
0.527 |
| 66 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.958 |
0.128 |
| 67 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.955 |
0.282 |
| 68 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.953 |
0.301 |
| 69 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.951 |
0.623 |
| 70 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.935 |
0.690 |
| 71 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.931 |
0.052 |
| 72 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.930 |
0.594 |
| 73 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.926 |
0.343 |
| 74 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.926 |
0.061 |
| 75 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.915 |
0.112 |
| 76 |
247913_at |
AT5G57510
|
unknown |
0.911 |
0.116 |
| 77 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.909 |
0.146 |
| 78 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.909 |
0.429 |
| 79 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.908 |
0.437 |
| 80 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.907 |
0.261 |
| 81 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.905 |
0.874 |
| 82 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.904 |
0.399 |
| 83 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.892 |
0.709 |
| 84 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.892 |
0.444 |
| 85 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.888 |
0.362 |
| 86 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.883 |
0.673 |
| 87 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.883 |
0.244 |
| 88 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
0.879 |
0.140 |
| 89 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.878 |
0.207 |
| 90 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.874 |
0.636 |
| 91 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.873 |
0.163 |
| 92 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.872 |
0.466 |
| 93 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.870 |
0.280 |
| 94 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.861 |
0.502 |
| 95 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.861 |
0.482 |
| 96 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.861 |
0.107 |
| 97 |
245755_at |
AT1G35210
|
unknown |
0.861 |
0.328 |
| 98 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.859 |
0.058 |
| 99 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.857 |
0.176 |
| 100 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.851 |
0.646 |
| 101 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.851 |
0.652 |
| 102 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.849 |
-0.250 |
| 103 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.840 |
0.417 |
| 104 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.839 |
0.062 |
| 105 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.839 |
0.033 |
| 106 |
247610_at |
AT5G60630
|
unknown |
0.838 |
0.627 |
| 107 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.833 |
0.456 |
| 108 |
260522_x_at |
AT2G41730
|
unknown |
0.831 |
0.013 |
| 109 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.822 |
0.472 |
| 110 |
253827_at |
AT4G28085
|
unknown |
0.820 |
0.087 |
| 111 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.818 |
0.487 |
| 112 |
256603_at |
AT3G28270
|
unknown |
-0.451 |
0.202 |
| 113 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.449 |
-0.160 |
| 114 |
250265_at |
AT5G12900
|
unknown |
-0.411 |
-0.068 |
| 115 |
261921_at |
AT1G65900
|
unknown |
-0.400 |
0.003 |
| 116 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.373 |
-0.110 |
| 117 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.371 |
0.044 |
| 118 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.362 |
-0.130 |
| 119 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.346 |
-0.090 |
| 120 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.343 |
-0.032 |
| 121 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.342 |
-0.227 |
| 122 |
266951_at |
AT2G18940
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.337 |
-0.117 |
| 123 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.328 |
-0.140 |
| 124 |
251907_at |
AT3G53760
|
ATGCP4, GCP4, GAMMA-TUBULIN COMPLEX PROTEIN 4 |
-0.324 |
-0.011 |
| 125 |
260395_at |
AT1G69780
|
ATHB13 |
-0.316 |
-0.105 |
| 126 |
257547_at |
AT3G13000
|
unknown |
-0.311 |
-0.099 |
| 127 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.310 |
-0.112 |
| 128 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.307 |
-0.084 |
| 129 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.303 |
-0.062 |
| 130 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.297 |
-0.005 |
| 131 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.296 |
-0.240 |
| 132 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.296 |
-0.073 |
| 133 |
261247_at |
AT1G20070
|
unknown |
-0.287 |
-0.003 |
| 134 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.282 |
-0.033 |
| 135 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.274 |
0.036 |
| 136 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.267 |
-0.014 |
| 137 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.267 |
-0.112 |
| 138 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
-0.266 |
-0.055 |
| 139 |
261609_at |
AT1G49740
|
[PLC-like phosphodiesterases superfamily protein] |
-0.265 |
-0.050 |
| 140 |
254764_at |
AT4G13250
|
NYC1, NON-YELLOW COLORING 1 |
-0.264 |
-0.028 |
| 141 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.262 |
-0.045 |
| 142 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-0.259 |
0.017 |
| 143 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.257 |
0.038 |
| 144 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.256 |
-0.138 |
| 145 |
254218_at |
AT4G23740
|
[Leucine-rich repeat protein kinase family protein] |
-0.253 |
-0.021 |
| 146 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.253 |
-0.089 |
| 147 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.252 |
-0.149 |
| 148 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.251 |
-0.143 |
| 149 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.251 |
-0.001 |
| 150 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.250 |
-0.128 |
| 151 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.249 |
-0.126 |
| 152 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.248 |
0.075 |
| 153 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.247 |
-0.061 |
| 154 |
250056_at |
AT5G17660
|
[tRNA (guanine-N-7) methyltransferase] |
-0.246 |
-0.114 |
| 155 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
-0.245 |
-0.064 |
| 156 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.244 |
-0.072 |
| 157 |
265986_at |
AT2G24230
|
[Leucine-rich repeat protein kinase family protein] |
-0.244 |
-0.031 |
| 158 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.243 |
-0.102 |
| 159 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.243 |
-0.005 |
| 160 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
-0.243 |
-0.069 |
| 161 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
-0.243 |
-0.143 |
| 162 |
251852_at |
AT3G54750
|
unknown |
-0.242 |
-0.048 |
| 163 |
259994_at |
AT1G68130
|
AtIDD14, indeterminate(ID)-domain 14, IDD14, indeterminate(ID)-domain 14, IDD14alpha, IDD14beta |
-0.242 |
-0.004 |
| 164 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.242 |
-0.131 |
| 165 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.240 |
-0.100 |
| 166 |
260989_at |
AT1G53450
|
unknown |
-0.236 |
-0.016 |
| 167 |
259775_at |
AT1G29530
|
unknown |
-0.236 |
-0.052 |
| 168 |
247098_at |
AT5G66470
|
[RNA binding] |
-0.235 |
-0.113 |
| 169 |
256899_at |
AT3G24660
|
TMKL1, transmembrane kinase-like 1 |
-0.233 |
-0.018 |
| 170 |
264911_at |
AT1G60690
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.232 |
-0.032 |
| 171 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
-0.231 |
-0.001 |
| 172 |
263793_at |
AT2G24630
|
ATCSLC08, ATCSLC8, CELLULOSE-SYNTHASE LIKE C8, CSLC08 |
-0.230 |
-0.082 |
| 173 |
258475_at |
AT3G02660
|
EMB2768, EMBRYO DEFECTIVE 2768, FAC31, EMBRYONIC FACTOR 31 |
-0.230 |
-0.121 |
| 174 |
255660_at |
AT4G00755
|
[F-box family protein] |
-0.230 |
-0.014 |
| 175 |
267578_at |
AT2G30695
|
unknown |
-0.229 |
-0.065 |
| 176 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
-0.228 |
-0.105 |
| 177 |
253574_at |
AT4G31030
|
[Putative membrane lipoprotein] |
-0.228 |
-0.139 |
| 178 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.226 |
-0.103 |
| 179 |
262907_at |
AT1G59720
|
CRR28, CHLORORESPIRATORY REDUCTION28 |
-0.226 |
-0.054 |
| 180 |
265990_at |
AT2G24280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.226 |
-0.058 |
| 181 |
266412_at |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
-0.225 |
0.052 |
| 182 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
-0.224 |
-0.154 |
| 183 |
245215_at |
AT1G67830
|
ATFXG1, Arabidopsis thaliana alpha-fucosidase 1, FXG1, alpha-fucosidase 1 |
-0.224 |
-0.136 |
| 184 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
-0.223 |
-0.160 |
| 185 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.223 |
-0.166 |
| 186 |
256706_at |
AT3G30300
|
[O-fucosyltransferase family protein] |
-0.222 |
-0.134 |
| 187 |
255785_at |
AT1G19920
|
APS2, ASA1, ATP SULFURYLASE ARABIDOPSIS 1 |
-0.221 |
-0.055 |
| 188 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.221 |
-0.060 |
| 189 |
245797_at |
AT1G45230
|
unknown |
-0.217 |
-0.115 |
| 190 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.216 |
-0.153 |
| 191 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.216 |
-0.024 |
| 192 |
251415_at |
AT3G60380
|
unknown |
-0.216 |
-0.124 |
| 193 |
261519_at |
AT1G71810
|
[Protein kinase superfamily protein] |
-0.215 |
-0.059 |
| 194 |
264517_at |
AT1G10120
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.214 |
-0.070 |
| 195 |
247417_at |
AT5G63040
|
unknown |
-0.213 |
-0.072 |
| 196 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.213 |
-0.075 |
| 197 |
251095_at |
AT5G01510
|
RUS5, ROOT UV-B SENSITIVE 5 |
-0.213 |
-0.155 |
| 198 |
248541_at |
AT5G50180
|
[Protein kinase superfamily protein] |
-0.213 |
-0.113 |
| 199 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.213 |
0.028 |
| 200 |
246520_at |
AT5G15790
|
[RING/U-box superfamily protein] |
-0.213 |
-0.004 |
| 201 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
-0.213 |
-0.150 |
| 202 |
257827_at |
AT3G26630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.212 |
-0.124 |
| 203 |
249282_at |
AT5G41760
|
[Nucleotide-sugar transporter family protein] |
-0.212 |
-0.182 |
| 204 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
-0.208 |
-0.119 |
| 205 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.206 |
-0.011 |
| 206 |
257773_at |
AT3G29185
|
unknown |
-0.204 |
-0.134 |
| 207 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.204 |
-0.068 |
| 208 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.203 |
-0.048 |
| 209 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.202 |
-0.104 |
| 210 |
248042_at |
AT5G55960
|
unknown |
-0.201 |
-0.107 |
| 211 |
253517_at |
AT4G31390
|
ABC1K1, ABC1-like kinase 1, ACDO1, ABC1-like kinase related to chlorophyll degradation and oxidative stress 1, AtACDO1, PGR6, PROTON GRADIENT REGULATION 6 |
-0.201 |
-0.050 |
| 212 |
258628_at |
AT3G02890
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.200 |
-0.062 |
| 213 |
258283_at |
AT3G26750
|
unknown |
-0.200 |
-0.069 |
| 214 |
264183_at |
AT1G65380
|
AtRLP10, Receptor Like Protein 10, CLV2, clavata 2 |
-0.198 |
-0.056 |
| 215 |
256343_at |
AT1G72090
|
[Methylthiotransferase] |
-0.198 |
-0.065 |
| 216 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.196 |
-0.189 |
| 217 |
264049_at |
AT2G22390
|
ATRABA4E, RABA4E, RAB GTPase homolog A4E |
-0.196 |
-0.043 |
| 218 |
250421_at |
AT5G11270
|
OCP3, overexpressor of cationic peroxidase 3 |
-0.196 |
-0.201 |
| 219 |
251343_at |
AT3G60620
|
CDS5, cytidinediphosphate diacylglycerol synthase 5 |
-0.196 |
-0.126 |
| 220 |
251193_at |
AT3G62910
|
APG3, ALBINO AND PALE GREEN, AtcpRF1, Arabidopsis thaliana chloroplast RF1, cpRF1, chloroplast ribosome release factor 1 |
-0.195 |
-0.065 |
| 221 |
246849_at |
AT5G26850
|
[Uncharacterized protein] |
-0.195 |
-0.113 |