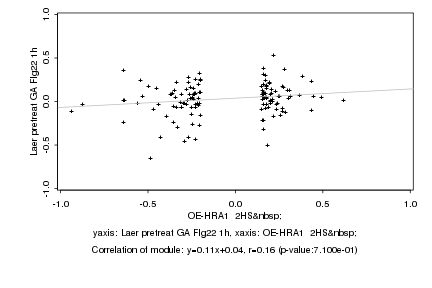
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
OE-HRA1 2HS |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.616 |
0.018 |
| 2 |
255527_at |
AT4G02360
|
unknown |
0.490 |
0.056 |
| 3 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.442 |
0.066 |
| 4 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.433 |
-0.099 |
| 5 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.430 |
0.233 |
| 6 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.383 |
0.288 |
| 7 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.364 |
0.080 |
| 8 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
0.310 |
0.058 |
| 9 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.309 |
0.135 |
| 10 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.298 |
0.036 |
| 11 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.296 |
0.137 |
| 12 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.286 |
-0.119 |
| 13 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.279 |
0.373 |
| 14 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.272 |
0.169 |
| 15 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
0.269 |
-0.070 |
| 16 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.269 |
0.180 |
| 17 |
263851_at |
AT2G04460
|
unknown |
0.265 |
-0.114 |
| 18 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
0.253 |
-0.156 |
| 19 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.252 |
0.060 |
| 20 |
248915_at |
AT5G45690
|
unknown |
0.243 |
0.062 |
| 21 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.236 |
-0.014 |
| 22 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.234 |
-0.032 |
| 23 |
261073_at |
AT1G07300
|
[josephin protein-related] |
0.234 |
-0.088 |
| 24 |
250821_at |
AT5G05190
|
unknown |
0.225 |
0.118 |
| 25 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.215 |
-0.172 |
| 26 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.213 |
0.534 |
| 27 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
0.208 |
0.004 |
| 28 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
0.207 |
0.031 |
| 29 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.203 |
0.140 |
| 30 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.201 |
0.096 |
| 31 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
0.198 |
0.090 |
| 32 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.197 |
0.020 |
| 33 |
252986_at |
AT4G38380
|
[MATE efflux family protein] |
0.196 |
0.005 |
| 34 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
0.195 |
-0.018 |
| 35 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.192 |
0.209 |
| 36 |
252400_at |
AT3G48020
|
unknown |
0.191 |
0.230 |
| 37 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.186 |
-0.068 |
| 38 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.179 |
-0.498 |
| 39 |
247177_at |
AT5G65300
|
unknown |
0.176 |
0.193 |
| 40 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.175 |
0.181 |
| 41 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
0.175 |
0.052 |
| 42 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.173 |
0.038 |
| 43 |
267604_at |
AT2G32930
|
ZFN2, zinc finger nuclease 2 |
0.173 |
0.152 |
| 44 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.173 |
-0.034 |
| 45 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.171 |
0.047 |
| 46 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.171 |
0.243 |
| 47 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
0.169 |
-0.079 |
| 48 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.168 |
0.302 |
| 49 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.167 |
0.094 |
| 50 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
0.166 |
0.042 |
| 51 |
248730_at |
AT5G48050
|
unknown |
0.163 |
0.040 |
| 52 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.163 |
0.093 |
| 53 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
0.160 |
0.205 |
| 54 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.159 |
0.382 |
| 55 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
0.159 |
-0.030 |
| 56 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.158 |
-0.207 |
| 57 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.158 |
-0.319 |
| 58 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.157 |
0.319 |
| 59 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
0.156 |
0.059 |
| 60 |
254479_at |
AT4G20350
|
[oxidoreductases] |
0.156 |
0.082 |
| 61 |
252675_at |
AT3G44270
|
[copia-like retrotransposon family, has a 1.1e-26 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.155 |
0.111 |
| 62 |
260631_at |
AT1G62350
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.155 |
0.089 |
| 63 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.155 |
0.099 |
| 64 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
0.152 |
-0.217 |
| 65 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.152 |
0.085 |
| 66 |
252773_at |
AT3G42910
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G07240.1)] |
0.152 |
0.089 |
| 67 |
258808_at |
AT3G04050
|
[Pyruvate kinase family protein] |
0.152 |
0.059 |
| 68 |
261551_at |
AT1G63440
|
HMA5, heavy metal atpase 5 |
0.151 |
0.133 |
| 69 |
257751_at |
AT3G18690
|
MKS1, MAP kinase substrate 1 |
0.151 |
0.034 |
| 70 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.149 |
0.182 |
| 71 |
247441_at |
AT5G62710
|
[Leucine-rich repeat protein kinase family protein] |
0.148 |
-0.090 |
| 72 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.942 |
-0.107 |
| 73 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.876 |
-0.026 |
| 74 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.644 |
0.016 |
| 75 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.642 |
0.367 |
| 76 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.641 |
-0.234 |
| 77 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.639 |
0.018 |
| 78 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
-0.562 |
-0.021 |
| 79 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.545 |
0.242 |
| 80 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.535 |
0.061 |
| 81 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.498 |
0.183 |
| 82 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.490 |
-0.654 |
| 83 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
-0.472 |
-0.085 |
| 84 |
260668_at |
AT1G19530
|
unknown |
-0.452 |
0.158 |
| 85 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.441 |
-0.029 |
| 86 |
259207_at |
AT3G09050
|
unknown |
-0.434 |
-0.412 |
| 87 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.395 |
-0.168 |
| 88 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.374 |
0.082 |
| 89 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.371 |
0.092 |
| 90 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.365 |
0.099 |
| 91 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
-0.359 |
-0.230 |
| 92 |
257375_at |
AT2G38640
|
unknown |
-0.356 |
-0.050 |
| 93 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.353 |
0.133 |
| 94 |
258091_at |
AT3G14560
|
unknown |
-0.345 |
0.052 |
| 95 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.341 |
-0.063 |
| 96 |
250211_at |
AT5G13880
|
unknown |
-0.339 |
0.221 |
| 97 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.336 |
-0.296 |
| 98 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.316 |
0.000 |
| 99 |
249384_at |
AT5G39890
|
unknown |
-0.311 |
0.082 |
| 100 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.310 |
-0.065 |
| 101 |
254200_at |
AT4G24110
|
unknown |
-0.309 |
-0.061 |
| 102 |
249211_at |
AT5G42680
|
unknown |
-0.299 |
-0.022 |
| 103 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.296 |
-0.457 |
| 104 |
255955_at |
AT1G22030
|
unknown |
-0.293 |
-0.019 |
| 105 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.282 |
0.144 |
| 106 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.280 |
-0.031 |
| 107 |
262741_at |
AT1G28760
|
[Uncharacterized conserved protein (DUF2215)] |
-0.276 |
0.023 |
| 108 |
250152_at |
AT5G15120
|
unknown |
-0.274 |
0.287 |
| 109 |
264800_at |
AT1G08800
|
MyoB1, myosin binding protein 1 |
-0.272 |
0.227 |
| 110 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.269 |
-0.409 |
| 111 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.265 |
0.081 |
| 112 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.262 |
0.039 |
| 113 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.258 |
0.172 |
| 114 |
252419_at |
AT3G47510
|
unknown |
-0.256 |
-0.140 |
| 115 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
-0.255 |
-0.061 |
| 116 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.249 |
0.054 |
| 117 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.249 |
0.045 |
| 118 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.248 |
-0.261 |
| 119 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
-0.245 |
0.089 |
| 120 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.245 |
0.158 |
| 121 |
265025_at |
AT1G24575
|
unknown |
-0.243 |
0.028 |
| 122 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.238 |
0.089 |
| 123 |
260395_at |
AT1G69780
|
ATHB13 |
-0.237 |
0.099 |
| 124 |
264277_at |
AT1G60390
|
PG1, polygalacturonase 1 |
-0.236 |
0.088 |
| 125 |
266880_at |
AT2G44770
|
[ELMO/CED-12 family protein] |
-0.234 |
0.100 |
| 126 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
-0.232 |
0.258 |
| 127 |
257140_at |
AT3G28910
|
ATMYB30, MYB30, myb domain protein 30 |
-0.231 |
-0.062 |
| 128 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.230 |
-0.429 |
| 129 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.230 |
-0.028 |
| 130 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.222 |
-0.013 |
| 131 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
-0.216 |
0.199 |
| 132 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.214 |
0.041 |
| 133 |
249762_at |
AT5G24000
|
unknown |
-0.213 |
-0.044 |
| 134 |
266956_at |
AT2G34510
|
unknown |
-0.209 |
-0.018 |
| 135 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.209 |
-0.273 |
| 136 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.208 |
0.115 |
| 137 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.206 |
0.325 |
| 138 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.205 |
0.242 |
| 139 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.204 |
0.111 |
| 140 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.202 |
-0.159 |
| 141 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.201 |
0.264 |