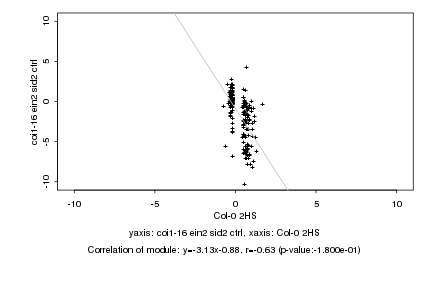
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 2HS |
Log2 signal ratio
coi1-16 ein2 sid2 ctrl |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.630 |
-0.274 |
| 2 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.287 |
-6.118 |
| 3 |
260668_at |
AT1G19530
|
unknown |
1.220 |
-4.457 |
| 4 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.167 |
-2.420 |
| 5 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
1.135 |
-1.861 |
| 6 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
1.069 |
-0.856 |
| 7 |
254200_at |
AT4G24110
|
unknown |
1.065 |
-7.368 |
| 8 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.043 |
-3.385 |
| 9 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
1.032 |
-4.334 |
| 10 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.014 |
-2.653 |
| 11 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
1.005 |
-8.116 |
| 12 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.992 |
-1.167 |
| 13 |
246001_at |
AT5G20790
|
unknown |
0.990 |
0.025 |
| 14 |
257925_at |
AT3G23170
|
unknown |
0.980 |
-0.759 |
| 15 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.948 |
-5.604 |
| 16 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.873 |
-7.847 |
| 17 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.870 |
-0.378 |
| 18 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.867 |
-6.528 |
| 19 |
261567_at |
AT1G33055
|
unknown |
0.860 |
-2.322 |
| 20 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.858 |
-0.715 |
| 21 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.833 |
-2.181 |
| 22 |
263182_at |
AT1G05575
|
unknown |
0.816 |
-6.620 |
| 23 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.806 |
-4.137 |
| 24 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.787 |
-3.470 |
| 25 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.773 |
-6.995 |
| 26 |
260005_at |
AT1G67920
|
unknown |
0.773 |
-5.447 |
| 27 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.770 |
-0.793 |
| 28 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.762 |
-1.692 |
| 29 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.751 |
-2.637 |
| 30 |
250472_at |
AT5G10210
|
unknown |
0.748 |
-0.891 |
| 31 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.747 |
-5.252 |
| 32 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.744 |
-5.764 |
| 33 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.738 |
-7.730 |
| 34 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.737 |
-6.706 |
| 35 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
0.725 |
-1.026 |
| 36 |
245041_at |
AT2G26530
|
AR781 |
0.725 |
-5.560 |
| 37 |
249384_at |
AT5G39890
|
unknown |
0.725 |
-1.872 |
| 38 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.718 |
-5.933 |
| 39 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.714 |
-1.524 |
| 40 |
247024_at |
AT5G66985
|
unknown |
0.701 |
-1.669 |
| 41 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.700 |
-3.386 |
| 42 |
259979_at |
AT1G76600
|
unknown |
0.697 |
-5.774 |
| 43 |
250152_at |
AT5G15120
|
unknown |
0.680 |
-0.635 |
| 44 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.677 |
-5.931 |
| 45 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.673 |
4.361 |
| 46 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.672 |
-1.555 |
| 47 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.671 |
-0.125 |
| 48 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.665 |
-6.144 |
| 49 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.663 |
-3.377 |
| 50 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.653 |
-0.530 |
| 51 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.649 |
-6.320 |
| 52 |
255884_at |
AT1G20310
|
unknown |
0.643 |
-6.002 |
| 53 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.643 |
-7.051 |
| 54 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.636 |
-6.581 |
| 55 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.624 |
-6.042 |
| 56 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.618 |
-2.313 |
| 57 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.601 |
-5.499 |
| 58 |
266658_at |
AT2G25735
|
unknown |
0.596 |
-7.002 |
| 59 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.596 |
-1.683 |
| 60 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.593 |
-6.331 |
| 61 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.592 |
-2.171 |
| 62 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.592 |
-1.741 |
| 63 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.583 |
-0.042 |
| 64 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.578 |
-0.317 |
| 65 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.572 |
-4.405 |
| 66 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.568 |
-2.386 |
| 67 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.564 |
1.432 |
| 68 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.564 |
-2.595 |
| 69 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
0.563 |
-2.647 |
| 70 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.555 |
-1.214 |
| 71 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.544 |
-1.317 |
| 72 |
253643_at |
AT4G29780
|
unknown |
0.541 |
-4.298 |
| 73 |
248432_at |
AT5G51390
|
unknown |
0.541 |
-0.395 |
| 74 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.540 |
-6.262 |
| 75 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.534 |
-4.161 |
| 76 |
264836_at |
AT1G03610
|
unknown |
0.527 |
0.018 |
| 77 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.519 |
-1.515 |
| 78 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.513 |
0.211 |
| 79 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.508 |
-10.219 |
| 80 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.503 |
-2.836 |
| 81 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.498 |
-6.365 |
| 82 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.497 |
-1.496 |
| 83 |
246584_at |
AT5G14730
|
unknown |
0.496 |
-4.039 |
| 84 |
259015_at |
AT3G07350
|
unknown |
0.494 |
-0.684 |
| 85 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.490 |
-2.306 |
| 86 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.487 |
-2.857 |
| 87 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.472 |
1.591 |
| 88 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.468 |
-5.005 |
| 89 |
265276_at |
AT2G28400
|
unknown |
0.467 |
-4.322 |
| 90 |
262741_at |
AT1G28760
|
[Uncharacterized conserved protein (DUF2215)] |
0.461 |
-0.137 |
| 91 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.459 |
-3.175 |
| 92 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.459 |
-3.989 |
| 93 |
258493_at |
AT3G02555
|
unknown |
0.457 |
-0.477 |
| 94 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.456 |
-0.565 |
| 95 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.455 |
-2.898 |
| 96 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.454 |
-4.018 |
| 97 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.453 |
-1.313 |
| 98 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.450 |
-2.951 |
| 99 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.447 |
0.513 |
| 100 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.446 |
0.603 |
| 101 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.441 |
-2.453 |
| 102 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.440 |
-5.975 |
| 103 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.437 |
-0.623 |
| 104 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.435 |
-4.463 |
| 105 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.792 |
-0.570 |
| 106 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.674 |
-5.588 |
| 107 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.535 |
2.230 |
| 108 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
-0.437 |
-0.192 |
| 109 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.429 |
0.089 |
| 110 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.397 |
0.706 |
| 111 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.376 |
1.229 |
| 112 |
264310_at |
AT1G62030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.365 |
1.054 |
| 113 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.364 |
-0.395 |
| 114 |
259298_at |
AT3G05370
|
AtRLP31, receptor like protein 31, RLP31, receptor like protein 31 |
-0.362 |
-1.656 |
| 115 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.342 |
0.176 |
| 116 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.338 |
0.319 |
| 117 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.337 |
-1.749 |
| 118 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.332 |
-1.448 |
| 119 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.330 |
1.390 |
| 120 |
255281_at |
AT4G04970
|
ATGSL01, GLUCAN SYNTHASE LIKE 1, ATGSL1, GLUCAN SYNTHASE LIKE-1, GSL01, GSL1, glucan synthase-like 1 |
-0.330 |
-0.048 |
| 121 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.329 |
0.956 |
| 122 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.328 |
1.143 |
| 123 |
263002_at |
AT1G54200
|
unknown |
-0.328 |
1.850 |
| 124 |
247141_at |
AT5G65560
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.320 |
-0.680 |
| 125 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.319 |
-0.066 |
| 126 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.319 |
1.327 |
| 127 |
259902_at |
AT1G74170
|
AtRLP13, receptor like protein 13, RLP13, receptor like protein 13 |
-0.319 |
-1.272 |
| 128 |
249184_at |
AT5G43020
|
[Leucine-rich repeat protein kinase family protein] |
-0.313 |
0.674 |
| 129 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.307 |
0.641 |
| 130 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.288 |
1.772 |
| 131 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.287 |
2.814 |
| 132 |
259689_x_at |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.285 |
0.364 |
| 133 |
262208_at |
AT1G74800
|
[Galactosyltransferase family protein] |
-0.285 |
0.352 |
| 134 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.284 |
1.286 |
| 135 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.280 |
0.263 |
| 136 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
-0.277 |
1.163 |
| 137 |
247701_at |
AT5G59900
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.275 |
0.440 |
| 138 |
251048_at |
AT5G02410
|
ALG10, homolog of yeast ALG10 |
-0.271 |
0.111 |
| 139 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
-0.269 |
0.260 |
| 140 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.264 |
1.019 |
| 141 |
247147_at |
AT5G65630
|
GTE7, global transcription factor group E7 |
-0.263 |
-1.365 |
| 142 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.261 |
1.801 |
| 143 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.260 |
0.947 |
| 144 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.259 |
0.519 |
| 145 |
253974_at |
AT4G26540
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.259 |
2.241 |
| 146 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.258 |
0.221 |
| 147 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.256 |
0.024 |
| 148 |
254043_at |
AT4G25990
|
CIL |
-0.256 |
0.069 |
| 149 |
247850_at |
AT5G58150
|
[Leucine-rich repeat protein kinase family protein] |
-0.256 |
0.099 |
| 150 |
246139_at |
AT5G19900
|
[PRLI-interacting factor, putative] |
-0.255 |
-0.497 |
| 151 |
261498_at |
AT1G28440
|
HSL1, HAESA-like 1 |
-0.253 |
1.034 |
| 152 |
247504_at |
AT5G61990
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.251 |
-0.600 |
| 153 |
265901_at |
AT2G25580
|
MEF8, mitochondrial editing factor 8 |
-0.251 |
-0.106 |
| 154 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.250 |
-0.485 |
| 155 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.243 |
-0.123 |
| 156 |
265039_at |
AT1G04000
|
unknown |
-0.239 |
0.395 |
| 157 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.238 |
1.625 |
| 158 |
253247_at |
AT4G34610
|
BLH6, BEL1-like homeodomain 6 |
-0.235 |
1.154 |
| 159 |
246893_at |
AT5G25520
|
[SPOC domain / Transcription elongation factor S-II protein] |
-0.233 |
0.139 |
| 160 |
248063_at |
AT5G55550
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.232 |
0.141 |
| 161 |
255151_at |
AT4G08180
|
ORP1C, OSBP(oxysterol binding protein)-related protein 1C |
-0.232 |
-2.017 |
| 162 |
245987_at |
AT5G13180
|
ANAC083, NAC domain containing protein 83, NAC083, NAC domain containing protein 83, VNI2, VND-interacting 2 |
-0.232 |
-0.345 |
| 163 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.231 |
0.933 |
| 164 |
248131_at |
AT5G54830
|
[DOMON domain-containing protein / dopamine beta-monooxygenase N-terminal domain-containing protein] |
-0.230 |
-1.021 |
| 165 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.230 |
0.184 |
| 166 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.229 |
0.603 |
| 167 |
252164_at |
AT3G50620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.229 |
-0.537 |
| 168 |
259605_at |
AT1G27910
|
ATPUB45, ARABIDOPSIS THALIANA PLANT U-BOX 45, PUB45, plant U-box 45 |
-0.228 |
0.040 |
| 169 |
248454_at |
AT5G51350
|
MOL1, MORE LATERAL GROWTH1 |
-0.226 |
0.440 |
| 170 |
251476_at |
AT3G59670
|
unknown |
-0.225 |
0.256 |
| 171 |
261750_at |
AT1G76120
|
[Pseudouridine synthase family protein] |
-0.225 |
0.378 |
| 172 |
261726_at |
AT1G76270
|
[O-fucosyltransferase family protein] |
-0.225 |
-0.711 |
| 173 |
254341_at |
AT4G22130
|
SRF8, STRUBBELIG-receptor family 8 |
-0.225 |
0.501 |
| 174 |
246964_at |
AT5G24830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.224 |
0.476 |
| 175 |
260718_at |
AT1G48110
|
ECT7, evolutionarily conserved C-terminal region 7 |
-0.224 |
-0.221 |
| 176 |
247952_at |
AT5G57250
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.222 |
0.226 |
| 177 |
256885_at |
AT3G15120
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.220 |
0.354 |
| 178 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
-0.218 |
0.833 |
| 179 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.216 |
0.624 |
| 180 |
250164_at |
AT5G15280
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.215 |
0.481 |
| 181 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.214 |
-0.002 |
| 182 |
256768_at |
AT3G13682
|
LDL2, LSD1-like2 |
-0.213 |
-0.156 |
| 183 |
254214_at |
AT4G23640
|
ATKT3, KUP4, TRH1, TINY ROOT HAIR 1 |
-0.213 |
-0.200 |
| 184 |
259191_at |
AT3G01720
|
unknown |
-0.213 |
-2.715 |
| 185 |
264183_at |
AT1G65380
|
AtRLP10, Receptor Like Protein 10, CLV2, clavata 2 |
-0.212 |
0.790 |
| 186 |
256157_at |
AT1G13580
|
LAG13, LAG1 longevity assurance homolog 3, LOH3, LAG One Homologue 3 |
-0.212 |
-0.071 |
| 187 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
-0.211 |
2.039 |
| 188 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
-0.210 |
-3.358 |
| 189 |
263042_at |
AT1G23340
|
unknown |
-0.208 |
2.228 |
| 190 |
266758_at |
AT2G46920
|
POL, poltergeist |
-0.206 |
-0.164 |
| 191 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.206 |
1.662 |
| 192 |
256622_at |
AT3G28920
|
AtHB34, homeobox protein 34, HB34, homeobox protein 34, ZHD9, ZINC FINGER HOMEODOMAIN 9 |
-0.205 |
0.584 |
| 193 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.205 |
-0.125 |
| 194 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.204 |
-0.127 |
| 195 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.203 |
-6.798 |
| 196 |
255428_at |
AT4G03390
|
SRF3, STRUBBELIG-receptor family 3 |
-0.203 |
0.241 |
| 197 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.202 |
1.250 |
| 198 |
251521_at |
AT3G59420
|
ACR4, crinkly4, CR4, crinkly4 |
-0.201 |
1.448 |
| 199 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
-0.201 |
-0.569 |
| 200 |
260081_at |
AT1G78170
|
unknown |
-0.201 |
1.767 |
| 201 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.200 |
0.453 |
| 202 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.200 |
-3.829 |
| 203 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
-0.199 |
-3.646 |
| 204 |
267221_at |
AT2G02480
|
STI, STICHEL |
-0.199 |
0.022 |
| 205 |
258704_at |
AT3G09780
|
ATCRR1, CCR1, CRINKLY4 related 1 |
-0.198 |
-0.325 |
| 206 |
263989_at |
AT2G42880
|
ATMPK20, MAP kinase 20, MPK20, MAP kinase 20 |
-0.196 |
0.345 |
| 207 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
-0.196 |
0.756 |