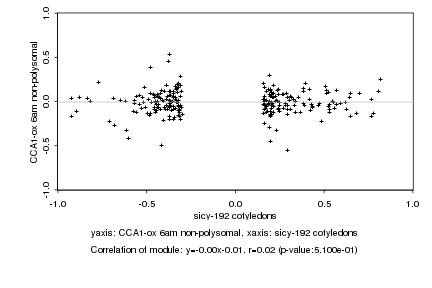
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sicy-192 cotyledons |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.813 |
0.257 |
| 2 |
251871_at |
AT3G54520
|
unknown |
0.802 |
0.122 |
| 3 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.775 |
-0.135 |
| 4 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.766 |
-0.167 |
| 5 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.763 |
0.023 |
| 6 |
252898_at |
AT4G39500
|
CYP96A11, cytochrome P450, family 96, subfamily A, polypeptide 11 |
0.695 |
0.094 |
| 7 |
252539_at |
AT3G45730
|
unknown |
0.681 |
-0.125 |
| 8 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.648 |
0.095 |
| 9 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.647 |
-0.161 |
| 10 |
255223_at |
AT4G05370
|
[BCS1 AAA-type ATPase] |
0.639 |
0.049 |
| 11 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.621 |
-0.081 |
| 12 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.619 |
-0.011 |
| 13 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.591 |
-0.018 |
| 14 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.569 |
0.128 |
| 15 |
254287_at |
AT4G22960
|
unknown |
0.565 |
-0.033 |
| 16 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.555 |
-0.071 |
| 17 |
256188_at |
AT1G30160
|
unknown |
0.548 |
-0.007 |
| 18 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.543 |
0.003 |
| 19 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.529 |
-0.090 |
| 20 |
256315_at |
AT1G35880
|
unknown |
0.528 |
-0.116 |
| 21 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.525 |
-0.023 |
| 22 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.522 |
-0.049 |
| 23 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.520 |
0.106 |
| 24 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.509 |
0.133 |
| 25 |
247772_at |
AT5G58610
|
[PHD finger transcription factor, putative] |
0.509 |
0.095 |
| 26 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.503 |
0.179 |
| 27 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.482 |
-0.224 |
| 28 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.472 |
-0.013 |
| 29 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.464 |
-0.043 |
| 30 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.433 |
-0.056 |
| 31 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.430 |
-0.059 |
| 32 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.423 |
-0.026 |
| 33 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.419 |
-0.097 |
| 34 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.417 |
0.030 |
| 35 |
265003_at |
AT1G26970
|
[Protein kinase superfamily protein] |
0.414 |
0.138 |
| 36 |
259918_at |
AT1G72570
|
[Integrase-type DNA-binding superfamily protein] |
0.392 |
0.205 |
| 37 |
253008_at |
AT4G38210
|
ATEXP20, ATEXPA20, expansin A20, ATHEXP ALPHA 1.23, EXP20, EXPANSIN 20, EXPA20, expansin A20 |
0.387 |
-0.042 |
| 38 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.380 |
0.153 |
| 39 |
260772_at |
AT1G49050
|
[Eukaryotic aspartyl protease family protein] |
0.380 |
0.116 |
| 40 |
261243_at |
AT1G20180
|
unknown |
0.379 |
-0.015 |
| 41 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.366 |
-0.123 |
| 42 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
0.352 |
0.056 |
| 43 |
263214_at |
AT1G30660
|
[nucleic acid binding] |
0.338 |
-0.120 |
| 44 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.335 |
0.003 |
| 45 |
253258_at |
AT4G34400
|
[AP2/B3-like transcriptional factor family protein] |
0.331 |
-0.039 |
| 46 |
251223_at |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
0.325 |
0.029 |
| 47 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.314 |
0.056 |
| 48 |
248938_at |
AT5G45780
|
[Leucine-rich repeat protein kinase family protein] |
0.309 |
-0.085 |
| 49 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.307 |
0.068 |
| 50 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.303 |
0.015 |
| 51 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
0.292 |
-0.085 |
| 52 |
259353_at |
AT3G05190
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.290 |
-0.546 |
| 53 |
253041_at |
AT4G37870
|
PCK1, phosphoenolpyruvate carboxykinase 1, PEPCK, PHOSPHOENOLPYRUVATE CARBOXYKINASE |
0.290 |
-0.045 |
| 54 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.289 |
0.047 |
| 55 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.288 |
-0.137 |
| 56 |
263730_at |
AT1G60090
|
BGLU4, beta glucosidase 4 |
0.286 |
0.094 |
| 57 |
250487_at |
AT5G09690
|
ATMGT7, ARABIDOPSIS THALIANA MAGNESIUM TRANSPORTER 7, MGT7, magnesium transporter 7, MRS2-7 |
0.278 |
-0.014 |
| 58 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
0.277 |
-0.040 |
| 59 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.270 |
-0.076 |
| 60 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.266 |
0.082 |
| 61 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.263 |
0.088 |
| 62 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.247 |
-0.044 |
| 63 |
264851_at |
AT2G17290
|
ATCDPK3, ARABIDOPSIS THALIANA CALMODULIN-DOMAIN PROTEIN KINASE 3, ATCPK6, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 6, CPK6, calcium dependent protein kinase 6 |
0.245 |
-0.011 |
| 64 |
250882_at |
AT5G04000
|
unknown |
0.243 |
-0.109 |
| 65 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
0.241 |
0.147 |
| 66 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
0.240 |
0.081 |
| 67 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.239 |
-0.088 |
| 68 |
247176_at |
AT5G65110
|
ACX2, acyl-CoA oxidase 2, ATACX2 |
0.238 |
-0.070 |
| 69 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.236 |
0.135 |
| 70 |
252990_at |
AT4G38440
|
IYO, MINIYO |
0.235 |
0.001 |
| 71 |
264946_at |
AT1G77010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.229 |
0.013 |
| 72 |
263690_at |
AT1G26960
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23 |
0.226 |
-0.327 |
| 73 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.223 |
0.058 |
| 74 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
0.223 |
0.007 |
| 75 |
245673_at |
AT1G56690
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.213 |
0.187 |
| 76 |
249178_at |
AT5G42890
|
ATSCP2, STEROL CARRIER PROTEIN 2, SCP2, sterol carrier protein 2 |
0.212 |
0.099 |
| 77 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.212 |
-0.123 |
| 78 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.210 |
0.054 |
| 79 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.210 |
-0.048 |
| 80 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.209 |
-0.049 |
| 81 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.207 |
-0.035 |
| 82 |
255590_at |
AT4G01610
|
[Cysteine proteinases superfamily protein] |
0.206 |
-0.015 |
| 83 |
258555_at |
AT3G06860
|
ATMFP2, MULTIFUNCTIONAL PROTEIN 2, MFP2, multifunctional protein 2 |
0.205 |
0.047 |
| 84 |
255318_at |
AT4G04190
|
unknown |
0.203 |
0.088 |
| 85 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.202 |
-0.075 |
| 86 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
0.202 |
0.112 |
| 87 |
264006_at |
AT2G22430
|
ATHB6, homeobox protein 6, HB6, homeobox protein 6 |
0.201 |
-0.145 |
| 88 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
0.199 |
-0.037 |
| 89 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.198 |
-0.009 |
| 90 |
246786_at |
AT5G27410
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.196 |
0.146 |
| 91 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.196 |
-0.165 |
| 92 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
0.195 |
-0.148 |
| 93 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.195 |
0.066 |
| 94 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
0.194 |
0.130 |
| 95 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.194 |
-0.140 |
| 96 |
247904_at |
AT5G57390
|
AIL5, AINTEGUMENTA-like 5, CHO1, CHOTTO 1, EMK, EMBRYOMAKER, PLT5, PLETHORA 5 |
0.193 |
-0.445 |
| 97 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.192 |
-0.112 |
| 98 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.189 |
0.083 |
| 99 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
0.189 |
0.002 |
| 100 |
257685_at |
AT3G12770
|
MEF22, mitochondrial editing factor 22 |
0.188 |
-0.284 |
| 101 |
264637_at |
AT1G65560
|
[Zinc-binding dehydrogenase family protein] |
0.188 |
0.302 |
| 102 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
0.185 |
-0.017 |
| 103 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.181 |
0.140 |
| 104 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.181 |
-0.029 |
| 105 |
255900_at |
AT1G17830
|
unknown |
0.176 |
-0.104 |
| 106 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
0.175 |
0.121 |
| 107 |
245670_at |
AT1G28210
|
ATJ1 |
0.173 |
-0.124 |
| 108 |
263151_at |
AT1G54120
|
unknown |
0.172 |
-0.053 |
| 109 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.170 |
0.015 |
| 110 |
252246_at |
AT3G49725
|
[GTP-binding protein, HflX] |
0.165 |
-0.034 |
| 111 |
264657_at |
AT1G09100
|
RPT5B, 26S proteasome AAA-ATPase subunit RPT5B |
0.164 |
-0.071 |
| 112 |
263986_at |
AT2G42790
|
CSY3, citrate synthase 3 |
0.163 |
0.026 |
| 113 |
247837_at |
AT5G57840
|
[HXXXD-type acyl-transferase family protein] |
0.161 |
0.067 |
| 114 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.161 |
-0.080 |
| 115 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.161 |
-0.037 |
| 116 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.160 |
0.161 |
| 117 |
263180_at |
AT1G05620
|
NSH2, nucleoside hydrolase 2, URH2, uridine-ribohydrolase 2 |
0.160 |
0.006 |
| 118 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
0.159 |
-0.238 |
| 119 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.159 |
-0.028 |
| 120 |
248426_at |
AT5G51740
|
[Peptidase family M48 family protein] |
0.159 |
-0.027 |
| 121 |
260028_at |
AT1G29980
|
unknown |
0.158 |
-0.130 |
| 122 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.156 |
0.209 |
| 123 |
248592_at |
AT5G49280
|
[hydroxyproline-rich glycoprotein family protein] |
0.154 |
-0.024 |
| 124 |
250029_at |
AT5G18200
|
[UTP:galactose-1-phosphate uridylyltransferases] |
0.153 |
0.026 |
| 125 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.928 |
-0.166 |
| 126 |
256096_at |
AT1G13650
|
unknown |
-0.927 |
0.043 |
| 127 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.896 |
-0.104 |
| 128 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.879 |
0.048 |
| 129 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.834 |
0.038 |
| 130 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.820 |
0.001 |
| 131 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.775 |
0.219 |
| 132 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.714 |
-0.218 |
| 133 |
246001_at |
AT5G20790
|
unknown |
-0.689 |
0.035 |
| 134 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.683 |
-0.265 |
| 135 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.652 |
0.012 |
| 136 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.622 |
0.009 |
| 137 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.614 |
-0.321 |
| 138 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.607 |
-0.415 |
| 139 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.575 |
-0.104 |
| 140 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.573 |
-0.013 |
| 141 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.571 |
0.012 |
| 142 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.563 |
-0.118 |
| 143 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.561 |
0.065 |
| 144 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.545 |
-0.028 |
| 145 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.541 |
0.071 |
| 146 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.533 |
-0.072 |
| 147 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.528 |
0.048 |
| 148 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.523 |
-0.006 |
| 149 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.514 |
0.167 |
| 150 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.508 |
-0.064 |
| 151 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.498 |
-0.133 |
| 152 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
-0.491 |
0.029 |
| 153 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.486 |
-0.157 |
| 154 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.484 |
0.097 |
| 155 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.483 |
-0.127 |
| 156 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.480 |
0.388 |
| 157 |
251036_at |
AT5G02160
|
unknown |
-0.477 |
-0.001 |
| 158 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.463 |
0.086 |
| 159 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.460 |
0.055 |
| 160 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.460 |
-0.059 |
| 161 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.459 |
0.070 |
| 162 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.457 |
-0.083 |
| 163 |
256446_at |
AT3G11110
|
[RING/U-box superfamily protein] |
-0.456 |
0.010 |
| 164 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.456 |
-0.003 |
| 165 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.453 |
-0.123 |
| 166 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.450 |
-0.026 |
| 167 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.447 |
-0.097 |
| 168 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.444 |
-0.063 |
| 169 |
265387_at |
AT2G20670
|
unknown |
-0.443 |
0.083 |
| 170 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.442 |
0.058 |
| 171 |
265611_at |
AT2G25510
|
unknown |
-0.442 |
0.007 |
| 172 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.439 |
0.050 |
| 173 |
263985_at |
AT2G42750
|
DJC77, DNA J protein C77 |
-0.439 |
-0.059 |
| 174 |
264096_at |
AT1G78995
|
unknown |
-0.438 |
-0.030 |
| 175 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.432 |
0.051 |
| 176 |
245007_at |
ATCG00350
|
PSAA |
-0.427 |
0.092 |
| 177 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.424 |
-0.081 |
| 178 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.423 |
0.044 |
| 179 |
260837_at |
AT1G43670
|
AtcFBP, Arabidopsis thaliana cytosolic fructose-1,6-bisphosphatase, FBP, fructose-1,6-bisphosphatase, FINS1, FRUCTOSE INSENSITIVE 1 |
-0.422 |
0.136 |
| 180 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.422 |
-0.493 |
| 181 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.413 |
0.012 |
| 182 |
258468_at |
AT3G06070
|
unknown |
-0.411 |
-0.207 |
| 183 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.407 |
0.002 |
| 184 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.405 |
-0.062 |
| 185 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-0.405 |
0.119 |
| 186 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.395 |
-0.082 |
| 187 |
247899_at |
AT5G57345
|
unknown |
-0.394 |
0.023 |
| 188 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.392 |
0.186 |
| 189 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.391 |
-0.034 |
| 190 |
261626_at |
AT1G01990
|
unknown |
-0.390 |
0.186 |
| 191 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.385 |
-0.088 |
| 192 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.384 |
0.066 |
| 193 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.382 |
0.456 |
| 194 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.376 |
-0.194 |
| 195 |
267063_at |
AT2G41120
|
unknown |
-0.376 |
-0.067 |
| 196 |
262566_at |
AT1G34310
|
ARF12, auxin response factor 12 |
-0.375 |
-0.015 |
| 197 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.375 |
0.076 |
| 198 |
260877_at |
AT1G21500
|
unknown |
-0.375 |
-0.051 |
| 199 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
-0.374 |
0.016 |
| 200 |
245351_at |
AT4G17640
|
CKB2, casein kinase II beta chain 2 |
-0.373 |
0.116 |
| 201 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.373 |
0.537 |
| 202 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
-0.372 |
-0.160 |
| 203 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.371 |
0.007 |
| 204 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.368 |
-0.041 |
| 205 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.367 |
0.115 |
| 206 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.367 |
0.071 |
| 207 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.363 |
-0.004 |
| 208 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.361 |
-0.096 |
| 209 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.355 |
0.006 |
| 210 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.354 |
0.096 |
| 211 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.354 |
-0.086 |
| 212 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.354 |
0.066 |
| 213 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.354 |
-0.194 |
| 214 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.353 |
0.047 |
| 215 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-0.352 |
0.117 |
| 216 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.350 |
-0.052 |
| 217 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.347 |
-0.174 |
| 218 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.342 |
0.056 |
| 219 |
265768_at |
AT2G48020
|
[Major facilitator superfamily protein] |
-0.340 |
-0.073 |
| 220 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.339 |
0.177 |
| 221 |
256228_at |
AT1G56190
|
[Phosphoglycerate kinase family protein] |
-0.339 |
0.030 |
| 222 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.335 |
0.142 |
| 223 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.333 |
-0.003 |
| 224 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.333 |
0.016 |
| 225 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.333 |
0.150 |
| 226 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.331 |
0.108 |
| 227 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.330 |
-0.006 |
| 228 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.327 |
0.200 |
| 229 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.326 |
-0.158 |
| 230 |
263943_at |
AT2G35800
|
SAMTL, S-adenosyl methionine transporter-like |
-0.326 |
-0.093 |
| 231 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.325 |
-0.038 |
| 232 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.324 |
0.208 |
| 233 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.322 |
0.163 |
| 234 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.321 |
0.018 |
| 235 |
246792_at |
AT5G27290
|
unknown |
-0.321 |
-0.051 |
| 236 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.318 |
-0.091 |
| 237 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
-0.318 |
-0.159 |
| 238 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.317 |
-0.114 |
| 239 |
253517_at |
AT4G31390
|
ABC1K1, ABC1-like kinase 1, ACDO1, ABC1-like kinase related to chlorophyll degradation and oxidative stress 1, AtACDO1, PGR6, PROTON GRADIENT REGULATION 6 |
-0.317 |
-0.088 |
| 240 |
250696_at |
AT5G06790
|
unknown |
-0.312 |
-0.034 |
| 241 |
249355_at |
AT5G40500
|
unknown |
-0.312 |
0.182 |
| 242 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.312 |
-0.201 |
| 243 |
249230_at |
AT5G42070
|
unknown |
-0.311 |
0.060 |
| 244 |
246158_at |
AT5G19855
|
AtRbcX2, RbcX2, homologue of cyanobacterial RbcX 2 |
-0.310 |
0.288 |
| 245 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
-0.309 |
-0.063 |
| 246 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.309 |
0.116 |
| 247 |
258929_at |
AT3G10060
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.303 |
-0.144 |