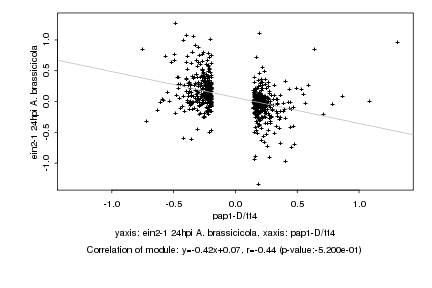
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pap1-D/tt4 |
Log2 signal ratio
ein2-1 24hpi A. brassicicola |
| 1 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
1.310 |
0.964 |
| 2 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
1.081 |
0.013 |
| 3 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.863 |
0.086 |
| 4 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.781 |
-0.044 |
| 5 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.707 |
-0.202 |
| 6 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.633 |
0.846 |
| 7 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.588 |
0.275 |
| 8 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.565 |
-0.020 |
| 9 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.543 |
0.122 |
| 10 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.529 |
0.203 |
| 11 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.489 |
0.222 |
| 12 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.470 |
-0.689 |
| 13 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.463 |
-0.396 |
| 14 |
256924_at |
AT3G29590
|
AT5MAT |
0.462 |
-0.097 |
| 15 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.451 |
-0.117 |
| 16 |
260688_at |
AT1G17665
|
unknown |
0.447 |
-0.741 |
| 17 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.439 |
-0.414 |
| 18 |
260668_at |
AT1G19530
|
unknown |
0.439 |
-0.002 |
| 19 |
264580_at |
AT1G05340
|
unknown |
0.436 |
-0.100 |
| 20 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.433 |
0.207 |
| 21 |
258383_at |
AT3G15440
|
unknown |
0.426 |
-0.002 |
| 22 |
253827_at |
AT4G28085
|
unknown |
0.403 |
-0.264 |
| 23 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.401 |
0.330 |
| 24 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.399 |
-0.965 |
| 25 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.391 |
-0.114 |
| 26 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.386 |
-0.026 |
| 27 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.383 |
-0.146 |
| 28 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.364 |
-0.250 |
| 29 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.362 |
0.106 |
| 30 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.358 |
-0.666 |
| 31 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.357 |
-0.159 |
| 32 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.356 |
-0.276 |
| 33 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.353 |
-0.438 |
| 34 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.340 |
-0.241 |
| 35 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.339 |
-0.023 |
| 36 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.337 |
-0.184 |
| 37 |
265954_at |
AT2G37260
|
ATWRKY44, DSL1, DR. STRANGELOVE 1, TTG2, TRANSPARENT TESTA GLABRA 2, WRKY44 |
0.337 |
-0.040 |
| 38 |
262910_at |
AT1G59710
|
unknown |
0.335 |
0.045 |
| 39 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.332 |
-0.403 |
| 40 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.328 |
0.044 |
| 41 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.326 |
-0.158 |
| 42 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.314 |
-0.516 |
| 43 |
257670_at |
AT3G20340
|
unknown |
0.311 |
0.270 |
| 44 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.308 |
0.103 |
| 45 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.303 |
-0.056 |
| 46 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.303 |
0.049 |
| 47 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
0.298 |
-0.139 |
| 48 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.296 |
-0.121 |
| 49 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.291 |
0.107 |
| 50 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
0.287 |
-0.246 |
| 51 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.285 |
-0.228 |
| 52 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
0.283 |
0.008 |
| 53 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.282 |
-0.347 |
| 54 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.280 |
-0.048 |
| 55 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
0.279 |
-0.084 |
| 56 |
257154_at |
AT3G27210
|
unknown |
0.278 |
-0.311 |
| 57 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
0.274 |
-0.894 |
| 58 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.274 |
-0.214 |
| 59 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.273 |
-0.275 |
| 60 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
0.272 |
0.168 |
| 61 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.271 |
-0.051 |
| 62 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.270 |
0.146 |
| 63 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.267 |
0.317 |
| 64 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.266 |
-0.467 |
| 65 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
0.264 |
-0.183 |
| 66 |
263222_at |
AT1G30640
|
[Protein kinase family protein] |
0.264 |
0.034 |
| 67 |
247305_at |
AT5G63905
|
unknown |
0.263 |
0.060 |
| 68 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
0.262 |
0.014 |
| 69 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.260 |
0.259 |
| 70 |
252217_at |
AT3G50140
|
unknown |
0.258 |
-0.341 |
| 71 |
258855_at |
AT3G02070
|
[Cysteine proteinases superfamily protein] |
0.257 |
-0.015 |
| 72 |
246664_at |
AT5G34800
|
[pseudogene] |
0.257 |
0.054 |
| 73 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.255 |
-0.126 |
| 74 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.255 |
-0.730 |
| 75 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.255 |
-0.049 |
| 76 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.252 |
0.079 |
| 77 |
251105_at |
AT5G01730
|
ATSCAR4, SCAR4, SCAR family protein 4, WAVE3 |
0.251 |
-0.075 |
| 78 |
264663_at |
AT1G09970
|
LRR XI-23, RLK7, receptor-like kinase 7 |
0.250 |
-0.017 |
| 79 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.248 |
0.218 |
| 80 |
252539_at |
AT3G45730
|
unknown |
0.248 |
-0.187 |
| 81 |
261039_at |
AT1G17455
|
ELF4-L4, ELF4-like 4 |
0.247 |
-0.524 |
| 82 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
0.247 |
-0.025 |
| 83 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.243 |
-0.158 |
| 84 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.243 |
0.024 |
| 85 |
250875_at |
AT5G04020
|
[calmodulin binding] |
0.240 |
-0.355 |
| 86 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.240 |
0.279 |
| 87 |
247394_at |
AT5G62865
|
unknown |
0.239 |
-0.189 |
| 88 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.237 |
0.364 |
| 89 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
0.237 |
-0.116 |
| 90 |
262103_at |
AT1G02940
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, GSTF5, glutathione S-transferase (class phi) 5 |
0.237 |
0.088 |
| 91 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.237 |
-0.257 |
| 92 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.236 |
-0.078 |
| 93 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.235 |
-0.068 |
| 94 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
0.235 |
-0.136 |
| 95 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.234 |
-0.116 |
| 96 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.234 |
0.490 |
| 97 |
260211_at |
AT1G74440
|
unknown |
0.233 |
-0.153 |
| 98 |
247878_at |
AT5G57760
|
unknown |
0.233 |
-0.652 |
| 99 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.231 |
-0.039 |
| 100 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.231 |
-0.051 |
| 101 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.230 |
0.084 |
| 102 |
267580_at |
AT2G41990
|
unknown |
0.230 |
-0.233 |
| 103 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.230 |
0.078 |
| 104 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.230 |
0.056 |
| 105 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
0.229 |
-0.004 |
| 106 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.226 |
0.030 |
| 107 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
0.224 |
-0.035 |
| 108 |
251934_at |
AT3G54070
|
[Ankyrin repeat family protein] |
0.221 |
-0.139 |
| 109 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.221 |
-0.556 |
| 110 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
0.217 |
0.099 |
| 111 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.216 |
0.213 |
| 112 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.216 |
0.310 |
| 113 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.216 |
-0.029 |
| 114 |
258608_at |
AT3G03020
|
unknown |
0.215 |
-0.143 |
| 115 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.214 |
-0.257 |
| 116 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.214 |
-0.160 |
| 117 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.214 |
0.265 |
| 118 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.213 |
0.564 |
| 119 |
253919_at |
AT4G27350
|
unknown |
0.213 |
0.024 |
| 120 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.213 |
-0.142 |
| 121 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.212 |
0.061 |
| 122 |
245266_at |
AT4G17070
|
[peptidyl-prolyl cis-trans isomerases] |
0.212 |
-0.073 |
| 123 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.211 |
-0.265 |
| 124 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
0.211 |
-0.003 |
| 125 |
257829_at |
AT3G26680
|
ATSNM1, SENSITIVE TO NITROGEN MUSTARD 1, SNM1, SENSITIVE TO NITROGEN MUSTARD 1 |
0.211 |
-0.085 |
| 126 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
0.210 |
-0.244 |
| 127 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.210 |
-0.037 |
| 128 |
263118_at |
AT1G03090
|
MCCA |
0.210 |
0.315 |
| 129 |
263919_at |
AT2G36470
|
unknown |
0.210 |
-0.621 |
| 130 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.210 |
0.216 |
| 131 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.209 |
0.190 |
| 132 |
256129_at |
AT1G18210
|
[Calcium-binding EF-hand family protein] |
0.208 |
-0.085 |
| 133 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.208 |
0.105 |
| 134 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
0.208 |
0.183 |
| 135 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.208 |
0.365 |
| 136 |
266017_at |
AT2G18690
|
unknown |
0.208 |
-0.067 |
| 137 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.207 |
-0.424 |
| 138 |
250301_at |
AT5G11970
|
unknown |
0.206 |
-0.112 |
| 139 |
259982_at |
AT1G76410
|
ATL8 |
0.206 |
-0.162 |
| 140 |
249987_at |
AT5G18490
|
unknown |
0.205 |
-0.109 |
| 141 |
257235_at |
AT3G15060
|
AtRABA1g, RAB GTPase homolog A1G, RABA1g, RAB GTPase homolog A1G |
0.204 |
-0.012 |
| 142 |
250480_at |
AT5G10290
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.204 |
0.052 |
| 143 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.203 |
-0.247 |
| 144 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.201 |
-0.137 |
| 145 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.201 |
0.302 |
| 146 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
0.200 |
-0.063 |
| 147 |
256848_at |
AT3G27960
|
KLCR2, kinesin light chain-related 2 |
0.200 |
0.141 |
| 148 |
256777_at |
AT3G13780
|
[SMAD/FHA domain-containing protein ] |
0.199 |
-0.048 |
| 149 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.199 |
-0.166 |
| 150 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
0.198 |
0.042 |
| 151 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.198 |
-0.305 |
| 152 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.197 |
0.036 |
| 153 |
261983_at |
AT1G33770
|
[Protein kinase superfamily protein] |
0.196 |
-0.084 |
| 154 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.196 |
0.267 |
| 155 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.195 |
-0.086 |
| 156 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.195 |
0.004 |
| 157 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.194 |
-0.002 |
| 158 |
252133_at |
AT3G50900
|
unknown |
0.194 |
0.074 |
| 159 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.193 |
0.017 |
| 160 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.192 |
-0.374 |
| 161 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.192 |
1.107 |
| 162 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.191 |
-0.183 |
| 163 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.191 |
0.048 |
| 164 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.191 |
0.031 |
| 165 |
249069_at |
AT5G44010
|
unknown |
0.191 |
-0.363 |
| 166 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.190 |
-0.295 |
| 167 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.189 |
-0.286 |
| 168 |
245166_at |
AT2G33170
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
0.188 |
-0.178 |
| 169 |
245566_at |
AT4G14610
|
[pseudogene] |
0.188 |
-0.174 |
| 170 |
248796_at |
AT5G47180
|
[Plant VAMP (vesicle-associated membrane protein) family protein] |
0.188 |
0.087 |
| 171 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
0.187 |
-0.069 |
| 172 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.187 |
-0.015 |
| 173 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.187 |
0.148 |
| 174 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.187 |
-0.087 |
| 175 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.187 |
0.456 |
| 176 |
247974_at |
AT5G56780
|
ATET2, ARABIDOPSIS EFFECTOR OF TRANSCRIPTION2, ET2, effector of transcription2 |
0.187 |
-0.044 |
| 177 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.187 |
-0.170 |
| 178 |
260641_at |
AT1G53200
|
unknown |
0.187 |
-0.033 |
| 179 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.186 |
-0.082 |
| 180 |
260939_at |
AT1G45180
|
[RING/U-box superfamily protein] |
0.186 |
0.017 |
| 181 |
251406_at |
AT3G60260
|
[ELMO/CED-12 family protein] |
0.186 |
-0.086 |
| 182 |
254408_at |
AT4G21390
|
B120 |
0.186 |
-0.105 |
| 183 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
0.185 |
-0.144 |
| 184 |
264390_at |
AT1G11950
|
[Transcription factor jumonji (jmjC) domain-containing protein] |
0.185 |
-0.138 |
| 185 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.185 |
-0.036 |
| 186 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
0.185 |
-0.275 |
| 187 |
256513_at |
AT1G31480
|
SGR2, SHOOT GRAVITROPISM 2 |
0.185 |
0.122 |
| 188 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.184 |
0.123 |
| 189 |
256069_at |
AT1G13740
|
AFP2, ABI five binding protein 2 |
0.184 |
-0.032 |
| 190 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.184 |
-0.148 |
| 191 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.184 |
-0.057 |
| 192 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.183 |
-0.398 |
| 193 |
254043_at |
AT4G25990
|
CIL |
0.183 |
-0.210 |
| 194 |
247412_at |
AT5G63010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.182 |
0.092 |
| 195 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.182 |
-0.098 |
| 196 |
260273_at |
AT1G80550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.181 |
0.017 |
| 197 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.181 |
0.355 |
| 198 |
251610_at |
AT3G57930
|
unknown |
0.180 |
-0.104 |
| 199 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
0.180 |
-0.056 |
| 200 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.180 |
0.253 |
| 201 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.180 |
-1.332 |
| 202 |
247835_at |
AT5G57910
|
unknown |
0.180 |
0.024 |
| 203 |
264841_at |
AT1G03740
|
[Protein kinase superfamily protein] |
0.179 |
-0.179 |
| 204 |
257621_at |
AT3G20410
|
CPK9, calmodulin-domain protein kinase 9 |
0.178 |
-0.140 |
| 205 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.178 |
-0.505 |
| 206 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.178 |
-0.002 |
| 207 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.178 |
0.125 |
| 208 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.178 |
0.225 |
| 209 |
257367_at |
AT2G25780
|
unknown |
0.178 |
-0.060 |
| 210 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.177 |
-0.240 |
| 211 |
264013_at |
AT2G21070
|
FIO1, FIONA1 |
0.176 |
-0.033 |
| 212 |
264852_at |
AT2G17480
|
ATMLO8, MILDEW RESISTANCE LOCUS O 8, MLO8, MILDEW RESISTANCE LOCUS O 8 |
0.175 |
0.047 |
| 213 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.175 |
0.019 |
| 214 |
258074_at |
AT3G25890
|
CRF11, cytokinin response factor 11 |
0.175 |
-0.030 |
| 215 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.175 |
0.085 |
| 216 |
253647_at |
AT4G29950
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.175 |
0.041 |
| 217 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.175 |
-0.061 |
| 218 |
267110_at |
AT2G14800
|
unknown |
0.174 |
-0.061 |
| 219 |
248869_at |
AT5G46840
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.173 |
0.064 |
| 220 |
259877_at |
AT1G76710
|
ASHH1, ASH1 RELATED PROTEIN 1, ASHH1, ASH1-RELATED PROTEIN 1, SDG26, SET domain group 26 |
0.172 |
-0.070 |
| 221 |
259876_at |
AT1G76700
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.172 |
-0.135 |
| 222 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
0.172 |
-0.098 |
| 223 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.172 |
-0.066 |
| 224 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.172 |
-0.348 |
| 225 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.172 |
-0.015 |
| 226 |
261823_at |
AT1G11400
|
PYM, partner of Y14-MAGO |
0.171 |
0.098 |
| 227 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
0.170 |
0.197 |
| 228 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.170 |
-0.460 |
| 229 |
253506_at |
AT4G31980
|
unknown |
0.169 |
-0.313 |
| 230 |
252602_at |
AT3G45040
|
[phosphatidate cytidylyltransferase family protein] |
0.169 |
-0.004 |
| 231 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
0.167 |
-0.052 |
| 232 |
267064_at |
AT2G41110
|
ATCAL5, CAM2, calmodulin 2 |
0.167 |
-0.019 |
| 233 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.167 |
-0.227 |
| 234 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
0.166 |
-0.034 |
| 235 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
0.166 |
0.025 |
| 236 |
246309_at |
AT3G51790
|
AtCCME, A. thaliana homolog of E. coli CcmE, ATG1, transmembrane protein G1P-related 1, G1, transmembrane protein G1P-related 1 |
0.165 |
-0.027 |
| 237 |
263076_at |
AT2G17520
|
ATIRE1-2, ARABIDOPSIS THALIANA INOSITOL REQUIRING 1-2, AtIRE1A, IRE1-2, INOSITOL REQUIRING 1-2, IRE1A |
0.165 |
-0.031 |
| 238 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.164 |
0.112 |
| 239 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.164 |
0.722 |
| 240 |
261181_at |
AT1G34580
|
[Major facilitator superfamily protein] |
0.164 |
0.052 |
| 241 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
0.164 |
-0.027 |
| 242 |
253322_at |
AT4G33980
|
unknown |
0.162 |
-0.891 |
| 243 |
257838_at |
AT3G25210
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.162 |
-0.182 |
| 244 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
0.162 |
-0.414 |
| 245 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
0.162 |
-0.124 |
| 246 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.162 |
-0.201 |
| 247 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.162 |
0.096 |
| 248 |
245405_at |
AT4G17150
|
[alpha/beta-Hydrolases superfamily protein] |
0.162 |
-0.202 |
| 249 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.161 |
0.098 |
| 250 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
0.161 |
-0.099 |
| 251 |
264238_at |
AT1G54740
|
unknown |
0.161 |
0.123 |
| 252 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
0.161 |
-0.157 |
| 253 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.161 |
-0.165 |
| 254 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.160 |
-0.129 |
| 255 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.160 |
-0.187 |
| 256 |
250776_at |
AT5G05320
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.160 |
-0.248 |
| 257 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
0.160 |
0.090 |
| 258 |
251086_at |
AT5G01450
|
APD2, ABERRANT POLLEN DEVELOPMENT 2 |
0.159 |
-0.032 |
| 259 |
265276_at |
AT2G28400
|
unknown |
0.159 |
0.165 |
| 260 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
0.159 |
-0.113 |
| 261 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.158 |
0.218 |
| 262 |
246529_at |
AT5G15730
|
AtCRLK2, CRLK2, calcium/calmodulin-regulated receptor-like kinase 2 |
0.157 |
0.014 |
| 263 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.156 |
-0.066 |
| 264 |
256413_at |
AT3G11100
|
[sequence-specific DNA binding transcription factors] |
0.156 |
0.014 |
| 265 |
262043_at |
AT1G80190
|
PSF1, partner of SLD five 1 |
0.155 |
-0.059 |
| 266 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
0.155 |
0.075 |
| 267 |
264423_at |
AT1G61690
|
[phosphoinositide binding] |
0.155 |
-0.088 |
| 268 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.155 |
0.103 |
| 269 |
264816_at |
AT1G03560
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.154 |
-0.031 |
| 270 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
0.154 |
-0.027 |
| 271 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.154 |
0.155 |
| 272 |
250555_at |
AT5G07950
|
unknown |
0.153 |
0.049 |
| 273 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
0.153 |
0.158 |
| 274 |
249947_at |
AT5G19200
|
TSC10B, TSC10B |
0.153 |
0.097 |
| 275 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.153 |
-0.004 |
| 276 |
260110_at |
AT1G63350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.152 |
-0.032 |
| 277 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.152 |
0.398 |
| 278 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.152 |
0.317 |
| 279 |
262845_at |
AT1G14740
|
TTA1, TITANIA 1 |
0.152 |
-0.039 |
| 280 |
262407_at |
AT1G34630
|
unknown |
0.152 |
-0.081 |
| 281 |
264447_at |
AT1G27300
|
unknown |
0.152 |
0.193 |
| 282 |
253075_at |
AT4G36150
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.152 |
0.001 |
| 283 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
0.152 |
0.156 |
| 284 |
260472_at |
AT1G10990
|
unknown |
0.151 |
0.007 |
| 285 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.150 |
0.070 |
| 286 |
253578_at |
AT4G30340
|
ATDGK7, diacylglycerol kinase 7, DGK7, diacylglycerol kinase 7 |
0.150 |
0.099 |
| 287 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
0.150 |
-0.181 |
| 288 |
249988_at |
AT5G18310
|
unknown |
0.149 |
0.045 |
| 289 |
254641_at |
AT4G18800
|
ATHSGBP, ATRAB11B, ATRABA1D, RAB GTPase homolog A1D, RABA1d, RAB GTPase homolog A1D |
0.149 |
-0.168 |
| 290 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.148 |
-0.931 |
| 291 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.148 |
-0.492 |
| 292 |
247845_at |
AT5G58090
|
[O-Glycosyl hydrolases family 17 protein] |
0.147 |
0.115 |
| 293 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
0.147 |
-0.015 |
| 294 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.147 |
0.145 |
| 295 |
249650_at |
AT5G37055
|
ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING |
0.146 |
-0.104 |
| 296 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.146 |
-0.106 |
| 297 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
0.146 |
0.194 |
| 298 |
257037_at |
AT3G19130
|
ATRBP47B, RNA-binding protein 47B, RBP47B, RNA-binding protein 47B |
0.146 |
-0.092 |
| 299 |
251630_at |
AT3G57420
|
unknown |
0.146 |
-0.014 |
| 300 |
255888_at |
AT1G20300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.145 |
0.021 |
| 301 |
256096_at |
AT1G13650
|
unknown |
-0.758 |
0.850 |
| 302 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.727 |
-0.321 |
| 303 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.635 |
-0.135 |
| 304 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.612 |
-0.002 |
| 305 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.594 |
0.049 |
| 306 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
-0.582 |
0.023 |
| 307 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.571 |
0.740 |
| 308 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.555 |
0.154 |
| 309 |
252661_at |
AT3G44450
|
unknown |
-0.536 |
0.013 |
| 310 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.521 |
0.638 |
| 311 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.500 |
0.769 |
| 312 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.497 |
0.673 |
| 313 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.492 |
-0.192 |
| 314 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.485 |
1.273 |
| 315 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.480 |
0.103 |
| 316 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.469 |
0.403 |
| 317 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.467 |
0.213 |
| 318 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.467 |
0.290 |
| 319 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.466 |
0.406 |
| 320 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.454 |
0.053 |
| 321 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.452 |
-0.098 |
| 322 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.450 |
0.288 |
| 323 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.433 |
-0.080 |
| 324 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.428 |
-0.598 |
| 325 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.424 |
1.001 |
| 326 |
251058_at |
AT5G01790
|
unknown |
-0.424 |
0.082 |
| 327 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.418 |
-0.153 |
| 328 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.414 |
0.271 |
| 329 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.398 |
1.084 |
| 330 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.398 |
0.169 |
| 331 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.398 |
0.649 |
| 332 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.397 |
0.368 |
| 333 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.396 |
0.377 |
| 334 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.394 |
0.522 |
| 335 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.394 |
0.735 |
| 336 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.388 |
0.022 |
| 337 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.388 |
-0.005 |
| 338 |
254683_at |
AT4G13800
|
unknown |
-0.385 |
0.276 |
| 339 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
-0.385 |
0.083 |
| 340 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.383 |
0.294 |
| 341 |
247751_at |
AT5G59050
|
unknown |
-0.382 |
0.132 |
| 342 |
252073_at |
AT3G51750
|
unknown |
-0.378 |
-0.019 |
| 343 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.378 |
0.305 |
| 344 |
254574_at |
AT4G19430
|
unknown |
-0.377 |
-0.125 |
| 345 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.374 |
0.312 |
| 346 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.373 |
0.165 |
| 347 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.369 |
0.001 |
| 348 |
261247_at |
AT1G20070
|
unknown |
-0.365 |
0.222 |
| 349 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.365 |
-0.060 |
| 350 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.364 |
0.268 |
| 351 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.364 |
0.292 |
| 352 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.361 |
-0.035 |
| 353 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.360 |
-0.609 |
| 354 |
254145_at |
AT4G24700
|
unknown |
-0.360 |
0.619 |
| 355 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.359 |
0.113 |
| 356 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.358 |
0.088 |
| 357 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.356 |
0.160 |
| 358 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.355 |
0.799 |
| 359 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.353 |
0.802 |
| 360 |
253044_at |
AT4G37290
|
unknown |
-0.351 |
-0.015 |
| 361 |
259275_at |
AT3G01060
|
unknown |
-0.350 |
0.422 |
| 362 |
257925_at |
AT3G23170
|
unknown |
-0.348 |
-0.118 |
| 363 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.345 |
-0.015 |
| 364 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.344 |
0.088 |
| 365 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.342 |
1.068 |
| 366 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.341 |
0.387 |
| 367 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.339 |
-0.102 |
| 368 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.336 |
0.266 |
| 369 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.335 |
0.398 |
| 370 |
253643_at |
AT4G29780
|
unknown |
-0.330 |
0.064 |
| 371 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.330 |
0.926 |
| 372 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.327 |
0.295 |
| 373 |
262211_at |
AT1G74930
|
ORA47 |
-0.327 |
0.193 |
| 374 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.321 |
0.450 |
| 375 |
261350_at |
AT1G79770
|
unknown |
-0.320 |
0.439 |
| 376 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.318 |
0.013 |
| 377 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.317 |
0.125 |
| 378 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.317 |
0.671 |
| 379 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.314 |
0.030 |
| 380 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.313 |
0.397 |
| 381 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.312 |
-0.003 |
| 382 |
258275_at |
AT3G15760
|
unknown |
-0.312 |
-0.133 |
| 383 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.311 |
0.086 |
| 384 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.310 |
0.131 |
| 385 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.310 |
0.570 |
| 386 |
264909_at |
AT2G17300
|
unknown |
-0.308 |
0.166 |
| 387 |
245692_at |
AT5G04150
|
BHLH101 |
-0.307 |
-0.449 |
| 388 |
247486_at |
AT5G62140
|
unknown |
-0.306 |
0.288 |
| 389 |
252010_at |
AT3G52740
|
unknown |
-0.304 |
0.323 |
| 390 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.304 |
0.251 |
| 391 |
265892_at |
AT2G15020
|
unknown |
-0.303 |
0.882 |
| 392 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.303 |
0.059 |
| 393 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.303 |
0.635 |
| 394 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
-0.303 |
0.150 |
| 395 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-0.303 |
0.034 |
| 396 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.301 |
0.260 |
| 397 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.298 |
-0.057 |
| 398 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.297 |
-0.013 |
| 399 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.297 |
-0.050 |
| 400 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.296 |
-0.053 |
| 401 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.294 |
-0.066 |
| 402 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.293 |
0.761 |
| 403 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.293 |
-0.015 |
| 404 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
-0.291 |
-0.195 |
| 405 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.290 |
0.054 |
| 406 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
-0.289 |
0.396 |
| 407 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.288 |
0.609 |
| 408 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.288 |
0.246 |
| 409 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.285 |
-0.054 |
| 410 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.284 |
-0.180 |
| 411 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.282 |
0.096 |
| 412 |
260696_at |
AT1G32520
|
unknown |
-0.281 |
0.421 |
| 413 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.280 |
0.186 |
| 414 |
259373_at |
AT1G69160
|
unknown |
-0.279 |
0.265 |
| 415 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.277 |
0.121 |
| 416 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.276 |
0.293 |
| 417 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.276 |
-0.037 |
| 418 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.275 |
0.195 |
| 419 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.273 |
0.417 |
| 420 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.272 |
0.512 |
| 421 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.272 |
0.774 |
| 422 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.271 |
0.030 |
| 423 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.270 |
0.351 |
| 424 |
247716_at |
AT5G59350
|
unknown |
-0.270 |
0.114 |
| 425 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.269 |
-0.040 |
| 426 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.269 |
0.525 |
| 427 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.269 |
0.203 |
| 428 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.268 |
0.526 |
| 429 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.268 |
0.661 |
| 430 |
256873_at |
AT3G26310
|
CYP71B35, cytochrome P450, family 71, subfamily B, polypeptide 35 |
-0.267 |
0.222 |
| 431 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.265 |
-0.015 |
| 432 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
-0.264 |
0.424 |
| 433 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.264 |
0.470 |
| 434 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
-0.263 |
0.414 |
| 435 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.263 |
-0.032 |
| 436 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.263 |
0.487 |
| 437 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.262 |
0.804 |
| 438 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.260 |
0.144 |
| 439 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.259 |
-0.246 |
| 440 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.259 |
0.222 |
| 441 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.259 |
0.336 |
| 442 |
246997_at |
AT5G67390
|
unknown |
-0.259 |
0.022 |
| 443 |
263829_at |
AT2G40435
|
unknown |
-0.258 |
0.408 |
| 444 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.256 |
0.083 |
| 445 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-0.256 |
-0.255 |
| 446 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.256 |
0.328 |
| 447 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.256 |
0.073 |
| 448 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.255 |
0.066 |
| 449 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.255 |
0.268 |
| 450 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.254 |
0.387 |
| 451 |
250828_at |
AT5G05250
|
unknown |
-0.254 |
-0.184 |
| 452 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.252 |
-0.243 |
| 453 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.252 |
0.383 |
| 454 |
266097_at |
AT2G37970
|
AtHBP2, HBP2, haem-binding protein 2, SOUL-1 |
-0.251 |
0.349 |
| 455 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.249 |
0.152 |
| 456 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.249 |
0.122 |
| 457 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.248 |
0.225 |
| 458 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.247 |
0.383 |
| 459 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.246 |
0.288 |
| 460 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.246 |
-0.008 |
| 461 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.246 |
0.374 |
| 462 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.246 |
0.135 |
| 463 |
252619_at |
AT3G45210
|
unknown |
-0.246 |
0.271 |
| 464 |
247047_at |
AT5G66650
|
unknown |
-0.245 |
0.055 |
| 465 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.245 |
0.339 |
| 466 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
-0.244 |
0.331 |
| 467 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.244 |
0.141 |
| 468 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.243 |
0.179 |
| 469 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.243 |
0.248 |
| 470 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.242 |
0.307 |
| 471 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.242 |
0.266 |
| 472 |
247977_at |
AT5G56850
|
unknown |
-0.241 |
0.253 |
| 473 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.240 |
0.120 |
| 474 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.238 |
-0.097 |
| 475 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.237 |
0.449 |
| 476 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.236 |
0.207 |
| 477 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.236 |
0.323 |
| 478 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.236 |
0.208 |
| 479 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.236 |
0.614 |
| 480 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.235 |
0.209 |
| 481 |
264201_at |
AT1G22630
|
unknown |
-0.235 |
0.070 |
| 482 |
246126_at |
AT5G20070
|
ATNUDT19, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 19, ATNUDX19, nudix hydrolase homolog 19, NUDX19, nudix hydrolase homolog 19 |
-0.235 |
0.302 |
| 483 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.235 |
0.034 |
| 484 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.233 |
0.671 |
| 485 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.233 |
0.179 |
| 486 |
256599_at |
AT3G14760
|
unknown |
-0.233 |
-0.147 |
| 487 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
-0.232 |
0.407 |
| 488 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.232 |
-0.133 |
| 489 |
265646_at |
AT2G27360
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.232 |
0.290 |
| 490 |
245743_at |
AT1G51080
|
unknown |
-0.231 |
-0.164 |
| 491 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.231 |
0.376 |
| 492 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.231 |
-0.153 |
| 493 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-0.230 |
-0.018 |
| 494 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.230 |
0.136 |
| 495 |
253228_at |
AT4G34630
|
unknown |
-0.229 |
0.082 |
| 496 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.228 |
0.336 |
| 497 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.228 |
0.327 |
| 498 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.228 |
0.194 |
| 499 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.228 |
0.168 |
| 500 |
259760_at |
AT1G77580
|
unknown |
-0.227 |
0.058 |
| 501 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.227 |
0.038 |
| 502 |
261661_at |
AT1G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.227 |
-0.111 |
| 503 |
251010_at |
AT5G02550
|
unknown |
-0.227 |
-0.001 |
| 504 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.226 |
0.054 |
| 505 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.225 |
0.135 |
| 506 |
261772_at |
AT1G76240
|
unknown |
-0.225 |
0.412 |
| 507 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.225 |
0.324 |
| 508 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
-0.224 |
0.793 |
| 509 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.224 |
0.266 |
| 510 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.223 |
0.238 |
| 511 |
267623_at |
AT2G39650
|
unknown |
-0.223 |
0.029 |
| 512 |
266337_at |
AT2G32390
|
ATGLR3.5, glutamate receptor 3.5, GLR3.5, glutamate receptor 3.5, GLR6 |
-0.223 |
-0.144 |
| 513 |
264636_at |
AT1G65490
|
unknown |
-0.222 |
0.579 |
| 514 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.222 |
0.222 |
| 515 |
248028_at |
AT5G55620
|
unknown |
-0.222 |
0.133 |
| 516 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.221 |
0.489 |
| 517 |
254791_at |
AT4G12910
|
scpl20, serine carboxypeptidase-like 20 |
-0.221 |
-0.086 |
| 518 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.221 |
0.391 |
| 519 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.221 |
0.152 |
| 520 |
264022_at |
AT2G21185
|
unknown |
-0.220 |
0.188 |
| 521 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.220 |
0.592 |
| 522 |
252888_at |
AT4G39210
|
APL3 |
-0.220 |
-0.198 |
| 523 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.218 |
0.334 |
| 524 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.218 |
0.114 |
| 525 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.218 |
0.095 |
| 526 |
261151_at |
AT1G19650
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.217 |
0.114 |
| 527 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.217 |
-0.491 |
| 528 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
-0.217 |
0.354 |
| 529 |
246531_at |
AT5G15800
|
AGL2, AGAMOUS-like 2, SEP1, SEPALLATA1 |
-0.217 |
-0.028 |
| 530 |
264840_at |
AT1G03440
|
[Leucine-rich repeat (LRR) family protein] |
-0.216 |
0.199 |
| 531 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.216 |
0.532 |
| 532 |
247324_at |
AT5G64190
|
unknown |
-0.216 |
0.051 |
| 533 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.215 |
0.180 |
| 534 |
246486_at |
AT5G15910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.215 |
0.278 |
| 535 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.215 |
0.542 |
| 536 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.214 |
0.227 |
| 537 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.214 |
-0.116 |
| 538 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.213 |
0.350 |
| 539 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.213 |
0.324 |
| 540 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.213 |
0.398 |
| 541 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.213 |
-0.083 |
| 542 |
259070_at |
AT3G11670
|
DGD1, DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1 |
-0.213 |
0.307 |
| 543 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.213 |
0.017 |
| 544 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.212 |
0.729 |
| 545 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.212 |
0.227 |
| 546 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.212 |
-0.020 |
| 547 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.212 |
-0.023 |
| 548 |
259460_at |
AT1G44000
|
unknown |
-0.211 |
0.318 |
| 549 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.210 |
0.071 |
| 550 |
248646_at |
AT5G49100
|
unknown |
-0.210 |
-0.049 |
| 551 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.210 |
0.188 |
| 552 |
258613_at |
AT3G02870
|
VTC4 |
-0.209 |
0.192 |
| 553 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-0.209 |
0.132 |
| 554 |
260515_at |
AT1G51460
|
ABCG13, ATP-binding cassette G13 |
-0.209 |
-0.014 |
| 555 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.208 |
0.281 |
| 556 |
249288_at |
AT5G41050
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.208 |
0.084 |
| 557 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.207 |
0.206 |
| 558 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.207 |
0.275 |
| 559 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.207 |
0.113 |
| 560 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.206 |
0.042 |
| 561 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.206 |
0.125 |
| 562 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.205 |
-0.150 |
| 563 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.204 |
0.245 |
| 564 |
259102_at |
AT3G11660
|
NHL1, NDR1/HIN1-like 1 |
-0.203 |
0.117 |
| 565 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.203 |
0.061 |
| 566 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.203 |
0.034 |
| 567 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.203 |
0.475 |
| 568 |
261921_at |
AT1G65900
|
unknown |
-0.203 |
-0.013 |
| 569 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.202 |
1.015 |
| 570 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.202 |
0.020 |
| 571 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.201 |
0.072 |
| 572 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.201 |
-0.014 |
| 573 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.201 |
0.050 |
| 574 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.201 |
-0.064 |
| 575 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.200 |
0.396 |
| 576 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.200 |
0.091 |
| 577 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.200 |
0.011 |
| 578 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.200 |
0.103 |
| 579 |
249303_at |
AT5G41460
|
unknown |
-0.199 |
0.045 |
| 580 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.199 |
0.216 |
| 581 |
253535_at |
AT4G31550
|
ATWRKY11, WRKY11, WRKY DNA-binding protein 11 |
-0.199 |
-0.080 |
| 582 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.199 |
0.317 |
| 583 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.198 |
0.199 |
| 584 |
263410_at |
AT2G04039
|
unknown |
-0.198 |
0.234 |
| 585 |
251727_at |
AT3G56290
|
unknown |
-0.197 |
0.621 |
| 586 |
261031_at |
AT1G17360
|
unknown |
-0.197 |
0.303 |
| 587 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.197 |
-0.065 |
| 588 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.197 |
-0.466 |
| 589 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.196 |
0.226 |
| 590 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
-0.196 |
0.237 |
| 591 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.196 |
0.087 |
| 592 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
-0.196 |
0.046 |
| 593 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.196 |
0.763 |
| 594 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.195 |
-0.047 |
| 595 |
247163_at |
AT5G65685
|
[UDP-Glycosyltransferase superfamily protein] |
-0.195 |
-0.007 |
| 596 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-0.195 |
0.286 |
| 597 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.195 |
0.435 |
| 598 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.194 |
-0.154 |
| 599 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.194 |
0.165 |
| 600 |
249091_at |
AT5G43860
|
ATCLH2, ARABIDOPSIS THALIANA CHLOROPHYLLASE 2, CLH2, chlorophyllase 2 |
-0.194 |
-0.029 |