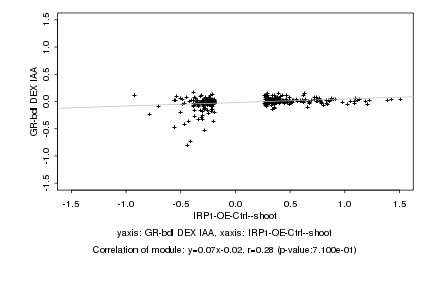
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IRP1-OE-Ctrl--shoot |
Log2 signal ratio
GR-bdl DEX IAA |
| 1 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
1.506 |
0.047 |
| 2 |
261684_at |
AT1G47400
|
unknown |
1.419 |
0.039 |
| 3 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
1.385 |
0.034 |
| 4 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.223 |
0.032 |
| 5 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.204 |
-0.047 |
| 6 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.189 |
0.015 |
| 7 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
1.130 |
0.038 |
| 8 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.108 |
0.026 |
| 9 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
1.093 |
0.056 |
| 10 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
1.081 |
-0.023 |
| 11 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
1.081 |
0.009 |
| 12 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.049 |
0.007 |
| 13 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.023 |
-0.052 |
| 14 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.973 |
-0.001 |
| 15 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.907 |
0.052 |
| 16 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.893 |
0.042 |
| 17 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.874 |
0.064 |
| 18 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.869 |
0.028 |
| 19 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.854 |
0.014 |
| 20 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.846 |
0.034 |
| 21 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.834 |
-0.041 |
| 22 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
0.830 |
-0.029 |
| 23 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.827 |
0.022 |
| 24 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.799 |
-0.062 |
| 25 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.782 |
-0.008 |
| 26 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.779 |
-0.026 |
| 27 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
0.774 |
0.001 |
| 28 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.771 |
0.031 |
| 29 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.768 |
0.057 |
| 30 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.748 |
0.005 |
| 31 |
258203_at |
AT3G13950
|
unknown |
0.743 |
0.038 |
| 32 |
244932_at |
ATCG01060
|
PSAC |
0.735 |
0.004 |
| 33 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.734 |
0.082 |
| 34 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.717 |
0.040 |
| 35 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.716 |
0.081 |
| 36 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.692 |
0.029 |
| 37 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.677 |
-0.032 |
| 38 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.671 |
-0.007 |
| 39 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
0.662 |
0.017 |
| 40 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.659 |
-0.107 |
| 41 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.639 |
0.050 |
| 42 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.624 |
0.152 |
| 43 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.615 |
0.113 |
| 44 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.615 |
0.009 |
| 45 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.615 |
-0.006 |
| 46 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.615 |
-0.016 |
| 47 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.613 |
0.007 |
| 48 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.611 |
0.024 |
| 49 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.607 |
0.025 |
| 50 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.592 |
0.020 |
| 51 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.590 |
0.003 |
| 52 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.559 |
0.006 |
| 53 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.557 |
0.052 |
| 54 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.523 |
0.027 |
| 55 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.521 |
-0.009 |
| 56 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.513 |
0.024 |
| 57 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.511 |
-0.014 |
| 58 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.499 |
0.014 |
| 59 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.495 |
-0.034 |
| 60 |
244929_at |
ATMG00580
|
NAD4, NADH dehydrogenase subunit 4 |
0.491 |
-0.045 |
| 61 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.490 |
0.035 |
| 62 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.490 |
0.089 |
| 63 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.488 |
-0.007 |
| 64 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
0.486 |
-0.041 |
| 65 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.470 |
-0.011 |
| 66 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.462 |
0.046 |
| 67 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
0.461 |
0.003 |
| 68 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.458 |
0.113 |
| 69 |
244962_at |
ATCG01050
|
NDHD |
0.454 |
0.004 |
| 70 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.453 |
0.035 |
| 71 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.449 |
-0.007 |
| 72 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
0.442 |
0.010 |
| 73 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.441 |
-0.024 |
| 74 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.439 |
-0.002 |
| 75 |
257323_at |
ATMG01200
|
ORF294 |
0.437 |
0.008 |
| 76 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
0.437 |
0.030 |
| 77 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.426 |
0.126 |
| 78 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.422 |
0.037 |
| 79 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.418 |
0.002 |
| 80 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.411 |
0.105 |
| 81 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.411 |
0.014 |
| 82 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.409 |
-0.001 |
| 83 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.408 |
-0.013 |
| 84 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.405 |
0.024 |
| 85 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
0.404 |
0.032 |
| 86 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.402 |
0.069 |
| 87 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.400 |
0.024 |
| 88 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.395 |
-0.030 |
| 89 |
265025_at |
AT1G24575
|
unknown |
0.391 |
0.037 |
| 90 |
257334_at |
ATMG01370
|
ORF111D |
0.389 |
0.032 |
| 91 |
264680_at |
AT1G65510
|
unknown |
0.386 |
0.005 |
| 92 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.386 |
0.162 |
| 93 |
246125_at |
AT5G19875
|
unknown |
0.380 |
-0.007 |
| 94 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.380 |
0.082 |
| 95 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.379 |
0.027 |
| 96 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.374 |
0.043 |
| 97 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.373 |
-0.020 |
| 98 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.373 |
0.013 |
| 99 |
244901_at |
ATMG00640
|
ORF25 |
0.369 |
0.008 |
| 100 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.369 |
0.047 |
| 101 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
0.368 |
0.057 |
| 102 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.367 |
0.070 |
| 103 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.367 |
-0.027 |
| 104 |
260754_at |
AT1G49000
|
unknown |
0.364 |
-0.009 |
| 105 |
260813_at |
AT1G43700
|
SUE3, sulphate utilization efficiency 3, VIP1, VIRE2-interacting protein 1 |
0.363 |
0.023 |
| 106 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.360 |
0.025 |
| 107 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.359 |
-0.021 |
| 108 |
245897_at |
AT5G09400
|
KUP7, K+ uptake permease 7 |
0.358 |
0.078 |
| 109 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.355 |
-0.100 |
| 110 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.353 |
-0.031 |
| 111 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.353 |
0.039 |
| 112 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.353 |
0.069 |
| 113 |
244933_at |
ATCG01070
|
NDHE |
0.352 |
-0.039 |
| 114 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.349 |
0.119 |
| 115 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.349 |
-0.116 |
| 116 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.342 |
0.018 |
| 117 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.341 |
0.005 |
| 118 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.338 |
0.047 |
| 119 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.337 |
-0.021 |
| 120 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.334 |
0.030 |
| 121 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.333 |
-0.133 |
| 122 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
0.333 |
0.124 |
| 123 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.331 |
0.051 |
| 124 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.331 |
0.046 |
| 125 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.329 |
-0.013 |
| 126 |
267178_at |
AT2G37750
|
unknown |
0.329 |
0.039 |
| 127 |
250909_at |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.329 |
-0.004 |
| 128 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.328 |
-0.030 |
| 129 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.324 |
0.025 |
| 130 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
0.324 |
-0.033 |
| 131 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
0.322 |
-0.002 |
| 132 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.322 |
0.038 |
| 133 |
261500_at |
AT1G28400
|
unknown |
0.321 |
-0.069 |
| 134 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.319 |
-0.015 |
| 135 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.318 |
0.052 |
| 136 |
258047_at |
AT3G21240
|
4CL2, 4-coumarate:CoA ligase 2, AT4CL2 |
0.317 |
0.047 |
| 137 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
0.314 |
0.044 |
| 138 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
0.313 |
0.052 |
| 139 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.311 |
0.025 |
| 140 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
0.310 |
-0.036 |
| 141 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.305 |
0.054 |
| 142 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.305 |
0.017 |
| 143 |
266808_at |
AT2G29995
|
unknown |
0.305 |
0.028 |
| 144 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
0.301 |
0.087 |
| 145 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.298 |
-0.023 |
| 146 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.295 |
0.023 |
| 147 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
0.292 |
0.027 |
| 148 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.291 |
-0.009 |
| 149 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.290 |
0.150 |
| 150 |
253277_at |
AT4G34230
|
ATCAD5, cinnamyl alcohol dehydrogenase 5, CAD-5, cinnamyl alcohol dehydrogenase 5, CAD5, cinnamyl alcohol dehydrogenase 5 |
0.290 |
0.034 |
| 151 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.290 |
-0.027 |
| 152 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.290 |
0.003 |
| 153 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.286 |
-0.017 |
| 154 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.286 |
0.113 |
| 155 |
265659_at |
AT2G25440
|
AtRLP20, receptor like protein 20, RLP20, receptor like protein 20 |
0.284 |
0.017 |
| 156 |
263769_at |
AT2G06390
|
transposable element gene |
0.284 |
-0.031 |
| 157 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
0.281 |
0.132 |
| 158 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.280 |
0.061 |
| 159 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.280 |
-0.033 |
| 160 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.279 |
0.094 |
| 161 |
249167_at |
AT5G42860
|
unknown |
0.278 |
0.007 |
| 162 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.277 |
0.042 |
| 163 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.277 |
-0.005 |
| 164 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.276 |
-0.064 |
| 165 |
251019_at |
AT5G02420
|
unknown |
0.276 |
-0.033 |
| 166 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.276 |
-0.083 |
| 167 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.274 |
0.091 |
| 168 |
245025_at |
ATCG00130
|
ATPF |
0.274 |
-0.043 |
| 169 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.273 |
0.003 |
| 170 |
248186_at |
AT5G53880
|
unknown |
0.270 |
-0.060 |
| 171 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.268 |
-0.005 |
| 172 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
0.268 |
0.017 |
| 173 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.267 |
0.016 |
| 174 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.267 |
0.048 |
| 175 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.265 |
0.117 |
| 176 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
0.265 |
-0.023 |
| 177 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.925 |
0.119 |
| 178 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.794 |
-0.221 |
| 179 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.709 |
-0.081 |
| 180 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.565 |
0.024 |
| 181 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.565 |
-0.467 |
| 182 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-0.559 |
0.037 |
| 183 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.553 |
0.037 |
| 184 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.548 |
0.102 |
| 185 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.511 |
-0.193 |
| 186 |
258356_at |
AT3G14340
|
unknown |
-0.503 |
0.066 |
| 187 |
253532_at |
AT4G31570
|
unknown |
-0.492 |
0.046 |
| 188 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
-0.485 |
-0.046 |
| 189 |
252963_at |
AT4G38820
|
unknown |
-0.472 |
-0.032 |
| 190 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.468 |
-0.411 |
| 191 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.452 |
0.088 |
| 192 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.442 |
-0.801 |
| 193 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.430 |
-0.353 |
| 194 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.428 |
0.011 |
| 195 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.413 |
-0.721 |
| 196 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.406 |
0.036 |
| 197 |
249824_at |
AT5G23380
|
unknown |
-0.389 |
0.025 |
| 198 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.389 |
0.179 |
| 199 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.386 |
-0.058 |
| 200 |
258204_at |
AT3G13960
|
AtGRF5, growth-regulating factor 5, GRF5, growth-regulating factor 5 |
-0.386 |
-0.090 |
| 201 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.386 |
-0.023 |
| 202 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.379 |
-0.161 |
| 203 |
252204_at |
AT3G50340
|
unknown |
-0.379 |
-0.259 |
| 204 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.378 |
0.074 |
| 205 |
263118_at |
AT1G03090
|
MCCA |
-0.368 |
0.059 |
| 206 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
-0.368 |
-0.049 |
| 207 |
245023_at |
ATCG00080
|
PSBI, photosystem II reaction center protein I |
-0.359 |
-0.021 |
| 208 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
-0.359 |
0.018 |
| 209 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.355 |
0.022 |
| 210 |
267397_at |
AT1G76170
|
[2-thiocytidine tRNA biosynthesis protein, TtcA] |
-0.353 |
-0.005 |
| 211 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.348 |
-0.037 |
| 212 |
267298_at |
AT2G23760
|
BLH4, BEL1-like homeodomain 4, SAW2, SAWTOOTH 2 |
-0.348 |
-0.031 |
| 213 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.346 |
-0.047 |
| 214 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.344 |
-0.082 |
| 215 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.342 |
-0.327 |
| 216 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.340 |
-0.058 |
| 217 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
-0.332 |
-0.000 |
| 218 |
247592_at |
AT5G60780
|
ATNRT2.3, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.3, NRT2.3, nitrate transporter 2.3 |
-0.325 |
-0.004 |
| 219 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.324 |
-0.157 |
| 220 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.320 |
0.104 |
| 221 |
252539_at |
AT3G45730
|
unknown |
-0.314 |
0.118 |
| 222 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.314 |
-0.022 |
| 223 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
-0.311 |
-0.280 |
| 224 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.311 |
-0.002 |
| 225 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
-0.308 |
-0.324 |
| 226 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.307 |
-0.035 |
| 227 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.307 |
-0.255 |
| 228 |
255959_at |
AT1G21980
|
ATPIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1, ATPIPK1, PIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1 |
-0.303 |
-0.170 |
| 229 |
266971_at |
AT2G39580
|
unknown |
-0.303 |
-0.050 |
| 230 |
266793_at |
AT2G03060
|
AGL30, AGAMOUS-like 30 |
-0.302 |
0.020 |
| 231 |
258501_at |
AT3G06780
|
[glycine-rich protein] |
-0.301 |
-0.025 |
| 232 |
244971_at |
ATCG00670
|
CLPP1, CASEINOLYTIC PROTEASE P 1, PCLPP, plastid-encoded CLP P |
-0.295 |
0.003 |
| 233 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
-0.294 |
0.032 |
| 234 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.294 |
0.022 |
| 235 |
259232_at |
AT3G11420
|
unknown |
-0.293 |
-0.089 |
| 236 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.293 |
-0.062 |
| 237 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.292 |
-0.135 |
| 238 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
-0.290 |
-0.015 |
| 239 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
-0.286 |
-0.111 |
| 240 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.286 |
-0.523 |
| 241 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.283 |
0.030 |
| 242 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.283 |
-0.036 |
| 243 |
249378_at |
AT5G40450
|
unknown |
-0.283 |
-0.030 |
| 244 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.282 |
-0.115 |
| 245 |
256813_at |
AT3G21360
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.279 |
0.090 |
| 246 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.279 |
0.067 |
| 247 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.277 |
0.014 |
| 248 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.275 |
-0.001 |
| 249 |
248874_at |
AT5G46460
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.275 |
-0.022 |
| 250 |
266749_at |
AT2G47060
|
PTI1-4, Pto-interacting 1-4 |
-0.273 |
-0.033 |
| 251 |
246385_at |
AT1G77390
|
CYCA1, CYCLIN A1, CYCA1;2, CYCLIN A1;2, DYP, TAM, TARDY ASYNCHRONOUS MEIOSIS |
-0.271 |
0.021 |
| 252 |
247885_at |
AT5G57830
|
unknown |
-0.271 |
-0.001 |
| 253 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.269 |
-0.014 |
| 254 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.265 |
-0.001 |
| 255 |
246796_at |
AT5G26770
|
unknown |
-0.265 |
0.046 |
| 256 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
-0.262 |
-0.048 |
| 257 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
-0.261 |
-0.162 |
| 258 |
255512_at |
AT4G02195
|
ATSYP42, ATTLG2B, SYP42, syntaxin of plants 42, TLG2B |
-0.261 |
0.013 |
| 259 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
-0.257 |
0.024 |
| 260 |
258855_at |
AT3G02070
|
[Cysteine proteinases superfamily protein] |
-0.257 |
-0.038 |
| 261 |
255422_at |
AT4G03270
|
CYCD6;1, Cyclin D6;1 |
-0.256 |
-0.066 |
| 262 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
-0.256 |
0.023 |
| 263 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
-0.256 |
0.017 |
| 264 |
246209_at |
AT4G36870
|
BLH2, BEL1-like homeodomain 2, SAW1, SAWTOOTH 1 |
-0.256 |
-0.063 |
| 265 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.254 |
-0.024 |
| 266 |
248585_at |
AT5G49640
|
unknown |
-0.252 |
0.059 |
| 267 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
-0.252 |
0.023 |
| 268 |
259651_at |
AT1G55280
|
[Lipase/lipooxygenase, PLAT/LH2 family protein] |
-0.252 |
0.010 |
| 269 |
245015_at |
ATCG00490
|
RBCL |
-0.250 |
-0.035 |
| 270 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
-0.249 |
0.025 |
| 271 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
-0.249 |
-0.211 |
| 272 |
251684_at |
AT3G56410
|
unknown |
-0.247 |
-0.037 |
| 273 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
-0.246 |
0.001 |
| 274 |
246211_at |
AT4G36730
|
AtGBF1, GBF1, G-box binding factor 1 |
-0.244 |
-0.001 |
| 275 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.244 |
0.036 |
| 276 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
-0.244 |
0.076 |
| 277 |
251380_at |
AT3G60700
|
unknown |
-0.243 |
0.024 |
| 278 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.243 |
-0.019 |
| 279 |
261451_at |
AT1G21060
|
unknown |
-0.242 |
-0.068 |
| 280 |
250569_at |
AT5G08130
|
BIM1 |
-0.241 |
0.005 |
| 281 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.240 |
0.039 |
| 282 |
262242_at |
AT1G48360
|
[zinc ion binding] |
-0.239 |
-0.047 |
| 283 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.239 |
-0.046 |
| 284 |
251310_at |
AT3G61150
|
HD-GL2-1, HOMEODOMAIN-GLABRA2 1, HDG1, homeodomain GLABROUS 1 |
-0.239 |
0.002 |
| 285 |
264238_at |
AT1G54740
|
unknown |
-0.237 |
0.124 |
| 286 |
244936_at |
ATCG01100
|
NDHA |
-0.236 |
-0.033 |
| 287 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.235 |
-0.007 |
| 288 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.234 |
0.071 |
| 289 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.234 |
0.014 |
| 290 |
257208_at |
AT3G14910
|
unknown |
-0.234 |
0.051 |
| 291 |
260054_at |
AT1G78130
|
UNE2, unfertilized embryo sac 2 |
-0.234 |
-0.040 |
| 292 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
-0.230 |
-0.014 |
| 293 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
-0.229 |
-0.018 |
| 294 |
265824_at |
AT2G35650
|
ATCSLA07, cellulose synthase like, ATCSLA7, CSLA07, CSLA07, cellulose synthase like, CSLA7, CELLULOSE SYNTHASE LIKE A7 |
-0.227 |
0.034 |
| 295 |
248784_at |
AT5G47380
|
unknown |
-0.227 |
-0.136 |
| 296 |
261863_at |
AT1G50630
|
unknown |
-0.226 |
0.019 |
| 297 |
259848_at |
AT1G72180
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.224 |
0.027 |
| 298 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.224 |
-0.041 |
| 299 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.224 |
-0.077 |
| 300 |
246053_at |
AT5G08340
|
[Nucleotidylyl transferase superfamily protein] |
-0.223 |
0.032 |
| 301 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
-0.223 |
-0.024 |
| 302 |
256958_at |
AT3G13430
|
[RING/U-box superfamily protein] |
-0.223 |
-0.029 |
| 303 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.221 |
0.006 |
| 304 |
255378_at |
AT4G03550
|
ATGSL05, glucan synthase-like 5, ATGSL5, EED3, enhancer of edr1 3, GSL05, GSL05, glucan synthase-like 5, GSL5, GLUCAN SYNTHASE-LIKE 5, PMR4, POWDERY MILDEW RESISTANT 4 |
-0.221 |
-0.051 |
| 305 |
260641_at |
AT1G53200
|
unknown |
-0.220 |
-0.043 |
| 306 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
-0.219 |
0.003 |
| 307 |
262407_at |
AT1G34630
|
unknown |
-0.219 |
0.011 |
| 308 |
249339_at |
AT5G41100
|
unknown |
-0.219 |
-0.034 |
| 309 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.219 |
-0.179 |
| 310 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.218 |
0.134 |
| 311 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.218 |
-0.081 |
| 312 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
-0.217 |
0.051 |
| 313 |
245926_at |
AT5G24740
|
SHBY, SHRUBBY |
-0.217 |
-0.018 |
| 314 |
255642_at |
AT4G00820
|
iqd17, IQ-domain 17 |
-0.216 |
-0.143 |
| 315 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.215 |
0.018 |
| 316 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
-0.215 |
-0.030 |
| 317 |
266380_at |
AT2G14680
|
MEE13, maternal effect embryo arrest 13 |
-0.214 |
-0.017 |
| 318 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.214 |
-0.036 |
| 319 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.214 |
0.027 |
| 320 |
253916_at |
AT4G27240
|
[zinc finger (C2H2 type) family protein] |
-0.212 |
-0.003 |
| 321 |
262627_at |
AT1G06580
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.211 |
-0.011 |
| 322 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.209 |
0.021 |
| 323 |
251225_at |
AT3G62660
|
GATL7, galacturonosyltransferase-like 7 |
-0.209 |
-0.046 |
| 324 |
257124_at |
AT3G20040
|
ATHXK4, HKL2, HEXOKINASE-LIKE 2 |
-0.209 |
0.016 |
| 325 |
257026_at |
AT3G19200
|
unknown |
-0.208 |
-0.354 |
| 326 |
257850_at |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
-0.208 |
-0.000 |
| 327 |
251544_at |
AT3G58790
|
GAUT15, galacturonosyltransferase 15 |
-0.208 |
-0.003 |
| 328 |
247127_at |
AT5G66100
|
AtLARP1b, LARP1b, La related protein 1b |
-0.208 |
0.002 |
| 329 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.208 |
0.017 |
| 330 |
261146_at |
AT1G19620
|
unknown |
-0.207 |
0.015 |
| 331 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
-0.207 |
0.013 |
| 332 |
260671_at |
AT1G19310
|
[RING/U-box superfamily protein] |
-0.207 |
-0.028 |
| 333 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
-0.206 |
-0.033 |
| 334 |
265088_at |
AT1G03750
|
CHR9, CHROMATIN REMODELING 9 |
-0.205 |
0.001 |
| 335 |
248913_at |
AT5G45760
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.205 |
-0.032 |
| 336 |
261087_at |
AT1G17350
|
[NADH:ubiquinone oxidoreductase intermediate-associated protein 30] |
-0.203 |
0.023 |
| 337 |
260160_at |
AT1G79880
|
AtLa2, La2, La protein 2 |
-0.201 |
-0.014 |
| 338 |
252408_at |
AT3G47600
|
ATMYB94, myb domain protein 94, ATMYBCP70, MYB94, myb domain protein 94 |
-0.200 |
-0.022 |
| 339 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.200 |
-0.018 |
| 340 |
263042_at |
AT1G23340
|
unknown |
-0.200 |
-0.200 |
| 341 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.199 |
0.013 |
| 342 |
251823_at |
AT3G55080
|
[SET domain-containing protein] |
-0.199 |
0.014 |
| 343 |
251558_at |
AT3G57810
|
[Cysteine proteinases superfamily protein] |
-0.199 |
0.048 |
| 344 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
-0.198 |
0.017 |
| 345 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.198 |
-0.003 |
| 346 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
-0.198 |
0.037 |
| 347 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
-0.197 |
0.003 |
| 348 |
249878_at |
AT5G23090
|
NF-YB13, nuclear factor Y, subunit B13 |
-0.197 |
-0.022 |
| 349 |
259230_at |
AT3G07780
|
OBE1, OBERON1 |
-0.196 |
-0.030 |
| 350 |
259072_at |
AT3G11700
|
FLA18, FASCICLIN-like arabinogalactan protein 18 precursor |
-0.196 |
-0.011 |
| 351 |
252113_at |
AT3G51620
|
[PAP/OAS1 substrate-binding domain superfamily] |
-0.195 |
-0.021 |