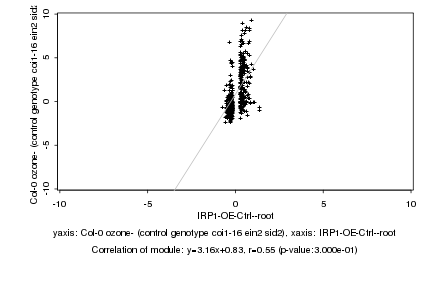
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IRP1-OE-Ctrl--root |
Log2 signal ratio
Col-0 ozone- (control genotype coi1-16 ein2 sid2) |
| 1 |
261684_at |
AT1G47400
|
unknown |
1.366 |
-0.659 |
| 2 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.324 |
-0.949 |
| 3 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
1.067 |
-0.041 |
| 4 |
253832_at |
AT4G27654
|
unknown |
1.025 |
-0.041 |
| 5 |
256442_at |
AT3G10930
|
unknown |
0.978 |
3.755 |
| 6 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.912 |
9.379 |
| 7 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.888 |
4.267 |
| 8 |
249893_at |
AT5G22555
|
unknown |
0.833 |
-0.041 |
| 9 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.824 |
2.841 |
| 10 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.807 |
2.892 |
| 11 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.802 |
-0.129 |
| 12 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.795 |
2.114 |
| 13 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.775 |
8.412 |
| 14 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.767 |
6.866 |
| 15 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.763 |
5.275 |
| 16 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.757 |
8.126 |
| 17 |
250098_at |
AT5G17350
|
unknown |
0.754 |
3.336 |
| 18 |
244962_at |
ATCG01050
|
NDHD |
0.747 |
0.433 |
| 19 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.725 |
6.665 |
| 20 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.723 |
0.820 |
| 21 |
253830_at |
AT4G27652
|
unknown |
0.719 |
2.197 |
| 22 |
249295_at |
AT5G41280
|
[Receptor-like protein kinase-related family protein] |
0.701 |
0.446 |
| 23 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.685 |
3.484 |
| 24 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.681 |
5.539 |
| 25 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.672 |
0.709 |
| 26 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.668 |
-1.509 |
| 27 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.650 |
3.917 |
| 28 |
253643_at |
AT4G29780
|
unknown |
0.650 |
3.881 |
| 29 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.640 |
1.827 |
| 30 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.636 |
2.821 |
| 31 |
260744_at |
AT1G15010
|
unknown |
0.623 |
2.290 |
| 32 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.622 |
-1.102 |
| 33 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.604 |
8.568 |
| 34 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
0.589 |
0.083 |
| 35 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.588 |
-0.049 |
| 36 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.586 |
-0.041 |
| 37 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.573 |
4.192 |
| 38 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.563 |
5.817 |
| 39 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.562 |
1.048 |
| 40 |
250747_at |
AT5G05900
|
[UDP-Glycosyltransferase superfamily protein] |
0.561 |
-0.041 |
| 41 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.535 |
-0.287 |
| 42 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.534 |
0.557 |
| 43 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.522 |
1.154 |
| 44 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.520 |
8.164 |
| 45 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.517 |
-0.041 |
| 46 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.515 |
4.715 |
| 47 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.514 |
4.781 |
| 48 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.498 |
4.834 |
| 49 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.494 |
7.853 |
| 50 |
245041_at |
AT2G26530
|
AR781 |
0.493 |
3.634 |
| 51 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.492 |
-0.041 |
| 52 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.490 |
3.940 |
| 53 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.484 |
3.267 |
| 54 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.483 |
2.263 |
| 55 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.483 |
0.464 |
| 56 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.480 |
1.462 |
| 57 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.477 |
4.305 |
| 58 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.475 |
1.434 |
| 59 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.470 |
1.270 |
| 60 |
244929_at |
ATMG00580
|
NAD4, NADH dehydrogenase subunit 4 |
0.468 |
-0.232 |
| 61 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.465 |
-1.038 |
| 62 |
246584_at |
AT5G14730
|
unknown |
0.463 |
4.028 |
| 63 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.460 |
4.905 |
| 64 |
254437_s_at |
AT3G06433
|
[pseudogene] |
0.456 |
-0.226 |
| 65 |
247177_at |
AT5G65300
|
unknown |
0.454 |
3.878 |
| 66 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
0.447 |
3.645 |
| 67 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
0.441 |
-0.498 |
| 68 |
245755_at |
AT1G35210
|
unknown |
0.439 |
4.993 |
| 69 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.438 |
4.848 |
| 70 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.438 |
-0.041 |
| 71 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.436 |
4.076 |
| 72 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.432 |
6.682 |
| 73 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.432 |
5.533 |
| 74 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.431 |
-0.258 |
| 75 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
0.431 |
-0.041 |
| 76 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.425 |
3.526 |
| 77 |
262211_at |
AT1G74930
|
ORA47 |
0.421 |
-0.936 |
| 78 |
245209_at |
AT5G12340
|
unknown |
0.419 |
5.056 |
| 79 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.417 |
5.074 |
| 80 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.414 |
5.223 |
| 81 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
0.413 |
-0.327 |
| 82 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.411 |
1.129 |
| 83 |
260642_at |
AT1G53260
|
[LOCATED IN: endomembrane system] |
0.407 |
0.508 |
| 84 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.405 |
4.528 |
| 85 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.402 |
4.960 |
| 86 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.402 |
6.978 |
| 87 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.402 |
4.449 |
| 88 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.401 |
-0.058 |
| 89 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.395 |
2.068 |
| 90 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.394 |
9.021 |
| 91 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
0.393 |
-0.581 |
| 92 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.392 |
4.115 |
| 93 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.391 |
0.640 |
| 94 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.390 |
3.002 |
| 95 |
263182_at |
AT1G05575
|
unknown |
0.387 |
4.579 |
| 96 |
260227_at |
AT1G74450
|
unknown |
0.385 |
0.838 |
| 97 |
258091_at |
AT3G14560
|
unknown |
0.384 |
0.322 |
| 98 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.383 |
8.211 |
| 99 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.383 |
3.210 |
| 100 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.380 |
1.911 |
| 101 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
0.380 |
-0.041 |
| 102 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.379 |
4.681 |
| 103 |
248717_at |
AT5G48175
|
unknown |
0.378 |
1.567 |
| 104 |
261690_at |
AT1G50090
|
AtBCAT7, BCAT7, branched-chain amino acid transaminase 7 |
0.377 |
-0.041 |
| 105 |
264861_at |
AT1G24320
|
[Six-hairpin glycosidases superfamily protein] |
0.374 |
-0.843 |
| 106 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
0.373 |
0.704 |
| 107 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.373 |
0.227 |
| 108 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.369 |
1.650 |
| 109 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.367 |
5.111 |
| 110 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.367 |
-0.069 |
| 111 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
0.365 |
-0.124 |
| 112 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
0.362 |
0.332 |
| 113 |
261193_at |
AT1G32920
|
unknown |
0.360 |
4.206 |
| 114 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.359 |
-0.041 |
| 115 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.359 |
5.041 |
| 116 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.356 |
3.444 |
| 117 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.355 |
-0.041 |
| 118 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.349 |
3.551 |
| 119 |
254178_at |
AT4G23880
|
unknown |
0.347 |
0.020 |
| 120 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.344 |
1.491 |
| 121 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.343 |
4.803 |
| 122 |
247708_at |
AT5G59550
|
unknown |
0.339 |
3.280 |
| 123 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.338 |
0.678 |
| 124 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.335 |
3.339 |
| 125 |
260813_at |
AT1G43700
|
SUE3, sulphate utilization efficiency 3, VIP1, VIRE2-interacting protein 1 |
0.332 |
0.594 |
| 126 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
0.330 |
0.253 |
| 127 |
263613_at |
AT2G25250
|
unknown |
0.330 |
1.622 |
| 128 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.329 |
5.038 |
| 129 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.327 |
3.462 |
| 130 |
245082_at |
AT2G23270
|
unknown |
0.326 |
7.624 |
| 131 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.325 |
4.835 |
| 132 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.323 |
6.556 |
| 133 |
260584_at |
AT2G43660
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.322 |
-1.288 |
| 134 |
265184_at |
AT1G23710
|
unknown |
0.321 |
3.263 |
| 135 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
0.321 |
-0.042 |
| 136 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.321 |
3.154 |
| 137 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.320 |
2.590 |
| 138 |
249072_at |
AT5G44060
|
unknown |
0.319 |
1.649 |
| 139 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.318 |
0.825 |
| 140 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.318 |
1.074 |
| 141 |
245381_at |
AT4G17785
|
MYB39, myb domain protein 39 |
0.317 |
-0.041 |
| 142 |
246270_at |
AT4G36500
|
unknown |
0.317 |
3.652 |
| 143 |
260012_at |
AT1G67865
|
unknown |
0.317 |
0.412 |
| 144 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.316 |
7.164 |
| 145 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
0.316 |
0.374 |
| 146 |
257334_at |
ATMG01370
|
ORF111D |
0.314 |
0.755 |
| 147 |
256609_at |
AT3G30843
|
unknown |
0.311 |
-0.041 |
| 148 |
260689_at |
AT1G32290
|
unknown |
0.310 |
-0.041 |
| 149 |
254091_at |
AT4G25070
|
unknown |
0.309 |
4.065 |
| 150 |
261405_at |
AT1G18740
|
unknown |
0.309 |
0.728 |
| 151 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.309 |
3.088 |
| 152 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.308 |
5.285 |
| 153 |
267623_at |
AT2G39650
|
unknown |
0.308 |
3.580 |
| 154 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.308 |
-0.924 |
| 155 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.307 |
3.475 |
| 156 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
0.306 |
1.843 |
| 157 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.305 |
3.822 |
| 158 |
245106_at |
AT2G41650
|
unknown |
0.305 |
-0.087 |
| 159 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.305 |
1.427 |
| 160 |
263569_at |
AT2G27170
|
SMC3, STRUCTURAL MAINTENANCE OF CHROMOSOMES 3, TTN7, TITAN7 |
0.305 |
0.005 |
| 161 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.305 |
-0.041 |
| 162 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.304 |
0.665 |
| 163 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.303 |
-0.033 |
| 164 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.303 |
1.637 |
| 165 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
0.302 |
-0.222 |
| 166 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.301 |
4.789 |
| 167 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.300 |
-0.799 |
| 168 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.299 |
5.308 |
| 169 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.299 |
0.875 |
| 170 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.298 |
1.404 |
| 171 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.296 |
2.482 |
| 172 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.295 |
5.122 |
| 173 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.294 |
-1.348 |
| 174 |
264635_at |
AT1G65500
|
unknown |
0.293 |
2.777 |
| 175 |
251410_at |
AT3G60280
|
UCC3, uclacyanin 3 |
0.293 |
-0.041 |
| 176 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.292 |
3.223 |
| 177 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
0.292 |
0.537 |
| 178 |
250676_at |
AT5G06320
|
NHL3, NDR1/HIN1-like 3 |
0.292 |
3.564 |
| 179 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.291 |
3.904 |
| 180 |
245677_at |
AT1G56660
|
unknown |
0.290 |
0.393 |
| 181 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.289 |
-0.213 |
| 182 |
265644_at |
AT2G27380
|
ATEPR1, extensin proline-rich 1, EPR1, extensin proline-rich 1 |
0.289 |
-0.796 |
| 183 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.285 |
1.572 |
| 184 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
0.285 |
-0.138 |
| 185 |
258973_at |
AT3G01900
|
CYP94B2, cytochrome P450, family 94, subfamily B, polypeptide 2 |
0.284 |
-0.416 |
| 186 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.284 |
-0.041 |
| 187 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.284 |
-0.605 |
| 188 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
0.283 |
0.875 |
| 189 |
248889_at |
AT5G46230
|
unknown |
0.282 |
1.518 |
| 190 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
0.282 |
0.027 |
| 191 |
266037_at |
AT2G05940
|
RIPK, RPM1-induced protein kinase |
0.281 |
2.153 |
| 192 |
257322_at |
ATMG01180
|
ORF111B |
0.281 |
-0.714 |
| 193 |
263931_at |
AT2G36220
|
unknown |
0.281 |
1.663 |
| 194 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.280 |
3.423 |
| 195 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
0.280 |
1.784 |
| 196 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.279 |
2.873 |
| 197 |
254364_at |
AT4G22020
|
[pseudogene] |
0.279 |
0.857 |
| 198 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.278 |
1.656 |
| 199 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.278 |
3.177 |
| 200 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.277 |
3.894 |
| 201 |
265194_at |
AT1G05010
|
ACO4, ethylene forming enzyme, EAT1, EFE, ethylene-forming enzyme |
0.277 |
3.776 |
| 202 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.276 |
4.398 |
| 203 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.276 |
-0.538 |
| 204 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.275 |
5.462 |
| 205 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.275 |
2.249 |
| 206 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.273 |
0.375 |
| 207 |
262017_at |
AT1G35550
|
[elongation factor Tu C-terminal domain-containing protein] |
0.272 |
-0.041 |
| 208 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.271 |
2.680 |
| 209 |
254213_at |
AT4G23590
|
[Tyrosine transaminase family protein] |
0.270 |
-1.172 |
| 210 |
251419_at |
AT3G60470
|
unknown |
0.270 |
2.180 |
| 211 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
0.270 |
1.626 |
| 212 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.269 |
-1.905 |
| 213 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.268 |
5.353 |
| 214 |
265618_at |
AT2G25460
|
unknown |
0.267 |
3.021 |
| 215 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
0.267 |
0.048 |
| 216 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
0.266 |
1.000 |
| 217 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
0.265 |
2.961 |
| 218 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.264 |
2.954 |
| 219 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.264 |
-0.041 |
| 220 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.263 |
-0.103 |
| 221 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.263 |
1.759 |
| 222 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.263 |
6.306 |
| 223 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
0.262 |
-0.041 |
| 224 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.260 |
4.777 |
| 225 |
251610_at |
AT3G57930
|
unknown |
0.259 |
-0.507 |
| 226 |
248072_at |
AT5G55680
|
[glycine-rich protein] |
0.258 |
0.237 |
| 227 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.258 |
1.100 |
| 228 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.257 |
6.938 |
| 229 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.257 |
4.602 |
| 230 |
266444_at |
AT2G43150
|
[Proline-rich extensin-like family protein] |
0.256 |
0.871 |
| 231 |
247913_at |
AT5G57510
|
unknown |
0.256 |
5.675 |
| 232 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.762 |
-0.641 |
| 233 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
-0.673 |
1.261 |
| 234 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.616 |
-2.322 |
| 235 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.580 |
-1.774 |
| 236 |
253532_at |
AT4G31570
|
unknown |
-0.562 |
0.012 |
| 237 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
-0.561 |
-0.191 |
| 238 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.543 |
-0.589 |
| 239 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.533 |
-1.685 |
| 240 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
-0.527 |
-0.756 |
| 241 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.516 |
-1.247 |
| 242 |
245023_at |
ATCG00080
|
PSBI, photosystem II reaction center protein I |
-0.514 |
0.230 |
| 243 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.514 |
1.836 |
| 244 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.497 |
-1.672 |
| 245 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.497 |
-1.326 |
| 246 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
-0.493 |
-0.346 |
| 247 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.483 |
0.520 |
| 248 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.479 |
-0.787 |
| 249 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.478 |
-1.253 |
| 250 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.475 |
-1.728 |
| 251 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.471 |
-1.834 |
| 252 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.468 |
-1.133 |
| 253 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
-0.468 |
-0.942 |
| 254 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.466 |
-1.039 |
| 255 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
-0.457 |
-0.979 |
| 256 |
256132_at |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.434 |
0.730 |
| 257 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.431 |
-0.147 |
| 258 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.429 |
-0.440 |
| 259 |
249482_at |
AT5G38980
|
unknown |
-0.424 |
-0.830 |
| 260 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.422 |
-0.547 |
| 261 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
-0.418 |
0.629 |
| 262 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
-0.411 |
-0.280 |
| 263 |
245234_at |
AT4G25560
|
AtMYB18, myb domain protein 18, LAF1, LONG AFTER FAR-RED LIGHT 1, MYB18, myb domain protein 18 |
-0.411 |
0.332 |
| 264 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.410 |
-1.574 |
| 265 |
245013_at |
ATCG00470
|
ATPE, ATP synthase epsilon chain |
-0.405 |
0.248 |
| 266 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.403 |
-1.638 |
| 267 |
261197_at |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
-0.391 |
-0.772 |
| 268 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.385 |
-1.457 |
| 269 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.382 |
-1.713 |
| 270 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-0.382 |
0.293 |
| 271 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.378 |
-1.892 |
| 272 |
261533_at |
AT1G71690
|
unknown |
-0.374 |
-1.106 |
| 273 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.373 |
1.294 |
| 274 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.371 |
1.964 |
| 275 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
-0.362 |
0.688 |
| 276 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.361 |
-0.566 |
| 277 |
266813_at |
AT2G44920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.359 |
-0.368 |
| 278 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.359 |
-0.393 |
| 279 |
245215_at |
AT1G67830
|
ATFXG1, Arabidopsis thaliana alpha-fucosidase 1, FXG1, alpha-fucosidase 1 |
-0.354 |
-0.453 |
| 280 |
259026_at |
AT3G09240
|
BSK9, brassinosteroid-signaling kinase 9 |
-0.353 |
-0.156 |
| 281 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-0.353 |
-0.041 |
| 282 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.353 |
-0.173 |
| 283 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.350 |
-1.730 |
| 284 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
-0.349 |
-0.542 |
| 285 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
-0.346 |
-0.712 |
| 286 |
267437_at |
AT2G19200
|
unknown |
-0.342 |
-0.041 |
| 287 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.341 |
0.317 |
| 288 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.339 |
-1.079 |
| 289 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.339 |
6.860 |
| 290 |
261460_at |
AT1G07880
|
ATMPK13 |
-0.339 |
0.717 |
| 291 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
-0.338 |
0.672 |
| 292 |
247320_at |
AT5G64040
|
PSAN |
-0.334 |
-0.587 |
| 293 |
259896_at |
AT1G71500
|
[Rieske (2Fe-2S) domain-containing protein] |
-0.333 |
-0.078 |
| 294 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.330 |
0.294 |
| 295 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.329 |
-1.661 |
| 296 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.328 |
-0.948 |
| 297 |
244971_at |
ATCG00670
|
CLPP1, CASEINOLYTIC PROTEASE P 1, PCLPP, plastid-encoded CLP P |
-0.325 |
-0.142 |
| 298 |
266551_at |
AT2G35260
|
unknown |
-0.325 |
-1.022 |
| 299 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.323 |
-0.633 |
| 300 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.323 |
-0.242 |
| 301 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
-0.322 |
-0.268 |
| 302 |
254263_at |
AT4G23493
|
unknown |
-0.321 |
-0.290 |
| 303 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.320 |
-0.381 |
| 304 |
260877_at |
AT1G21500
|
unknown |
-0.316 |
-0.551 |
| 305 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
-0.313 |
1.629 |
| 306 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.312 |
-0.922 |
| 307 |
252130_at |
AT3G50820
|
OEC33, OXYGEN EVOLVING COMPLEX SUBUNIT 33 KDA, PSBO-2, PHOTOSYSTEM II SUBUNIT O-2, PSBO2, photosystem II subunit O-2 |
-0.311 |
-0.804 |
| 308 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.311 |
-2.178 |
| 309 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.310 |
0.500 |
| 310 |
263946_at |
AT2G36000
|
EMB3114, EMBRYO DEFECTIVE 3114 |
-0.310 |
-1.314 |
| 311 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.309 |
-1.380 |
| 312 |
251365_at |
AT3G61310
|
AHL11, AT-hook motif nuclear localized protein 11 |
-0.307 |
-0.482 |
| 313 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.307 |
-1.156 |
| 314 |
252517_at |
AT3G46340
|
[Leucine-rich repeat protein kinase family protein] |
-0.306 |
4.800 |
| 315 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
-0.306 |
-0.421 |
| 316 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.305 |
-0.519 |
| 317 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.304 |
-2.164 |
| 318 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
-0.304 |
-0.427 |
| 319 |
257966_at |
AT3G19800
|
unknown |
-0.303 |
-1.008 |
| 320 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
-0.302 |
-0.041 |
| 321 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.302 |
-0.942 |
| 322 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
-0.301 |
-1.139 |
| 323 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
-0.300 |
-0.044 |
| 324 |
251878_at |
AT3G54310
|
unknown |
-0.300 |
-0.044 |
| 325 |
255957_at |
AT1G22160
|
unknown |
-0.298 |
-1.433 |
| 326 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.295 |
1.937 |
| 327 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.295 |
-0.135 |
| 328 |
251036_at |
AT5G02160
|
unknown |
-0.294 |
-0.776 |
| 329 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.294 |
2.367 |
| 330 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.291 |
-2.367 |
| 331 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.289 |
-0.112 |
| 332 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.289 |
-0.978 |
| 333 |
263002_at |
AT1G54200
|
unknown |
-0.287 |
-1.665 |
| 334 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.285 |
-0.292 |
| 335 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.284 |
3.070 |
| 336 |
262632_at |
AT1G06680
|
OE23, OXYGEN EVOLVING COMPLEX SUBUNIT 23 KDA, OEE2, OXYGEN-EVOLVING ENHANCER PROTEIN 2, PSBP-1, photosystem II subunit P-1, PSII-P, PHOTOSYSTEM II SUBUNIT P |
-0.283 |
-0.590 |
| 337 |
255264_at |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.281 |
-1.252 |
| 338 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.280 |
-0.603 |
| 339 |
258939_at |
AT3G10020
|
unknown |
-0.277 |
-0.267 |
| 340 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.275 |
1.067 |
| 341 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.275 |
-0.041 |
| 342 |
253067_at |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
-0.274 |
0.332 |
| 343 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.274 |
-0.818 |
| 344 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.274 |
-1.486 |
| 345 |
248491_at |
AT5G51010
|
[Rubredoxin-like superfamily protein] |
-0.273 |
-0.434 |
| 346 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.272 |
0.748 |
| 347 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.271 |
0.581 |
| 348 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.271 |
2.496 |
| 349 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.269 |
-1.278 |
| 350 |
251243_at |
AT3G61870
|
unknown |
-0.267 |
-0.284 |
| 351 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
-0.265 |
-0.652 |
| 352 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.265 |
-0.041 |
| 353 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.265 |
-1.552 |
| 354 |
261921_at |
AT1G65900
|
unknown |
-0.265 |
-1.191 |
| 355 |
247895_at |
AT5G58010
|
LRL3, LJRHL1-like 3 |
-0.265 |
-0.041 |
| 356 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.264 |
-1.942 |
| 357 |
265309_at |
AT2G20290
|
ATXIG, MYOSIN XI G, XIG, myosin-like protein XIG |
-0.262 |
-0.813 |
| 358 |
247807_at |
AT5G58360
|
ATOFP3, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 3, OFP3, ovate family protein 3 |
-0.261 |
0.261 |
| 359 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.260 |
1.035 |
| 360 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.260 |
-0.939 |
| 361 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
-0.259 |
-0.411 |
| 362 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.259 |
-0.754 |
| 363 |
245806_at |
AT1G45474
|
Lhca5, photosystem I light harvesting complex gene 5 |
-0.258 |
-0.332 |
| 364 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.257 |
4.527 |
| 365 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.257 |
-0.041 |
| 366 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.256 |
0.535 |
| 367 |
247813_at |
AT5G58330
|
[lactate/malate dehydrogenase family protein] |
-0.254 |
-0.513 |
| 368 |
250856_at |
AT5G04810
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.254 |
-1.220 |
| 369 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.254 |
0.568 |
| 370 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
-0.254 |
4.365 |
| 371 |
255285_at |
AT4G04630
|
unknown |
-0.254 |
-0.670 |
| 372 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.253 |
-0.041 |
| 373 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.253 |
0.793 |
| 374 |
245263_at |
AT4G17740
|
[Peptidase S41 family protein] |
-0.252 |
-0.371 |
| 375 |
262558_at |
AT1G31335
|
unknown |
-0.251 |
0.401 |
| 376 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.251 |
-0.750 |
| 377 |
253865_at |
AT4G27470
|
ATRMA3, RMA3, RING membrane-anchor 3 |
-0.251 |
0.569 |
| 378 |
245006_at |
ATCG00340
|
PSAB |
-0.251 |
0.177 |
| 379 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.250 |
-1.511 |
| 380 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.250 |
0.003 |
| 381 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
-0.249 |
-1.122 |
| 382 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.248 |
0.812 |
| 383 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.247 |
-0.781 |
| 384 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.247 |
0.060 |
| 385 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.247 |
-0.730 |
| 386 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.246 |
-0.583 |
| 387 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
-0.245 |
0.754 |
| 388 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
-0.244 |
-1.350 |
| 389 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.244 |
-0.640 |
| 390 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.243 |
0.226 |
| 391 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.243 |
-0.635 |
| 392 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.241 |
-1.777 |
| 393 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.241 |
-0.428 |
| 394 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.239 |
-0.558 |
| 395 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.239 |
-1.145 |
| 396 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.239 |
-0.547 |
| 397 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.237 |
-0.534 |
| 398 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.235 |
-0.341 |
| 399 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.235 |
-1.588 |
| 400 |
248287_at |
AT5G52970
|
[thylakoid lumen 15.0 kDa protein] |
-0.235 |
-0.456 |
| 401 |
250190_at |
AT5G14320
|
EMB3137, EMBRYO DEFECTIVE 3137 |
-0.234 |
-0.197 |
| 402 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.234 |
-0.028 |
| 403 |
262331_at |
AT1G64050
|
unknown |
-0.234 |
-0.009 |
| 404 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.234 |
-0.552 |
| 405 |
247688_at |
AT5G59760
|
unknown |
-0.233 |
-0.392 |
| 406 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.233 |
0.285 |
| 407 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-0.233 |
1.000 |
| 408 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.232 |
-0.587 |
| 409 |
255290_at |
AT4G04640
|
ATPC1 |
-0.232 |
-0.602 |
| 410 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.232 |
-1.142 |
| 411 |
258643_at |
AT3G08010
|
ATAB2 |
-0.231 |
-1.055 |
| 412 |
245713_at |
AT5G04370
|
NAMT1 |
-0.229 |
0.629 |
| 413 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
-0.229 |
1.828 |
| 414 |
260759_at |
AT1G49180
|
[protein kinase family protein] |
-0.228 |
-0.374 |
| 415 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.228 |
-0.170 |
| 416 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.228 |
-0.586 |
| 417 |
252036_at |
AT3G52070
|
unknown |
-0.228 |
-1.427 |
| 418 |
251646_at |
AT3G57780
|
unknown |
-0.227 |
-0.329 |
| 419 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.227 |
0.267 |
| 420 |
246617_at |
AT5G36270
|
[similar to DHAR2, glutathione dehydrogenase (ascorbate) [Arabidopsis thaliana] (TAIR:AT1G75270.1)] |
-0.227 |
-0.041 |
| 421 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.227 |
-0.462 |
| 422 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.226 |
0.425 |
| 423 |
250160_at |
AT5G15210
|
ATHB30, homeobox protein 30, HB30, homeobox protein 30, ZFHD3, ZINC FINGER HOMEODOMAIN 3, ZHD8, ZINC FINGER HOMEODOMAIN 8 |
-0.226 |
-0.551 |
| 424 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
-0.226 |
-0.561 |
| 425 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
-0.224 |
-0.490 |
| 426 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.224 |
0.774 |
| 427 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.223 |
-1.024 |
| 428 |
261952_at |
AT1G64430
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.223 |
-0.388 |
| 429 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.223 |
-0.625 |
| 430 |
265539_at |
AT2G15830
|
unknown |
-0.223 |
1.515 |
| 431 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.222 |
-1.662 |
| 432 |
264352_at |
AT1G03270
|
unknown |
-0.222 |
-1.111 |
| 433 |
250535_at |
AT5G08480
|
[VQ motif-containing protein] |
-0.222 |
-0.555 |
| 434 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.221 |
-0.863 |
| 435 |
249824_at |
AT5G23380
|
unknown |
-0.221 |
0.063 |
| 436 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.220 |
-0.735 |
| 437 |
259195_at |
AT3G01730
|
unknown |
-0.220 |
-0.041 |
| 438 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
-0.220 |
1.311 |
| 439 |
259340_at |
AT3G03870
|
unknown |
-0.220 |
0.701 |
| 440 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.219 |
-0.753 |
| 441 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.219 |
-0.407 |
| 442 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.219 |
4.034 |
| 443 |
257647_at |
AT3G25805
|
unknown |
-0.219 |
-0.354 |
| 444 |
246054_at |
AT5G08360
|
unknown |
-0.219 |
0.574 |
| 445 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.219 |
-1.842 |
| 446 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
-0.219 |
-0.041 |
| 447 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.218 |
1.946 |
| 448 |
246616_at |
AT5G36260
|
[Eukaryotic aspartyl protease family protein] |
-0.218 |
4.521 |
| 449 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.217 |
-0.902 |
| 450 |
255766_at |
AT1G16750
|
unknown |
-0.216 |
-0.589 |
| 451 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.216 |
-1.001 |
| 452 |
247231_at |
AT5G65230
|
AtMYB53, myb domain protein 53, MYB53, myb domain protein 53 |
-0.215 |
-0.188 |
| 453 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
-0.214 |
1.363 |
| 454 |
248585_at |
AT5G49640
|
unknown |
-0.213 |
-0.060 |
| 455 |
254105_at |
AT4G25080
|
CHLM, magnesium-protoporphyrin IX methyltransferase |
-0.212 |
-0.960 |
| 456 |
256813_at |
AT3G21360
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.212 |
-1.198 |
| 457 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.212 |
-0.739 |
| 458 |
249922_at |
AT5G19140
|
AILP1, ATAILP1 |
-0.211 |
-0.728 |
| 459 |
257132_at |
AT3G20230
|
[Ribosomal L18p/L5e family protein] |
-0.211 |
-0.694 |
| 460 |
250654_at |
AT5G06940
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.210 |
-0.302 |
| 461 |
258250_at |
AT3G15850
|
ADS3, AtADS3, FAD5, fatty acid desaturase 5, FADB, FATTY ACID DESATURASE B, JB67 |
-0.210 |
-1.315 |