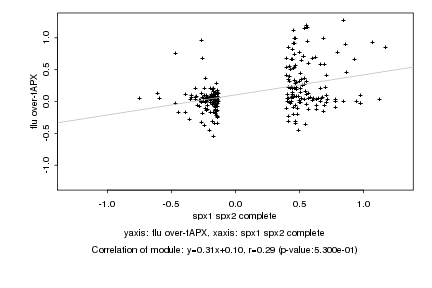
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
spx1 spx2 complete |
Log2 signal ratio
flu over-tAPX |
| 1 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.171 |
0.853 |
| 2 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
1.120 |
0.040 |
| 3 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.065 |
0.929 |
| 4 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.976 |
-0.020 |
| 5 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.972 |
0.109 |
| 6 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.970 |
-0.016 |
| 7 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.945 |
0.003 |
| 8 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.925 |
0.669 |
| 9 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.867 |
0.468 |
| 10 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.857 |
0.901 |
| 11 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.844 |
1.287 |
| 12 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.841 |
0.016 |
| 13 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.790 |
0.780 |
| 14 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.782 |
-0.093 |
| 15 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.778 |
0.044 |
| 16 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.775 |
0.011 |
| 17 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
0.716 |
0.044 |
| 18 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.711 |
0.421 |
| 19 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.708 |
0.100 |
| 20 |
263851_at |
AT2G04460
|
unknown |
0.706 |
-0.001 |
| 21 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.698 |
0.230 |
| 22 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.696 |
0.590 |
| 23 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.694 |
-0.055 |
| 24 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.687 |
1.004 |
| 25 |
246001_at |
AT5G20790
|
unknown |
0.684 |
-0.144 |
| 26 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.677 |
0.078 |
| 27 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.672 |
0.063 |
| 28 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.664 |
0.589 |
| 29 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.660 |
0.218 |
| 30 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.658 |
0.014 |
| 31 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.649 |
0.053 |
| 32 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.633 |
-0.049 |
| 33 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.630 |
-0.123 |
| 34 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.627 |
0.042 |
| 35 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.620 |
0.693 |
| 36 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.605 |
0.063 |
| 37 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.601 |
0.684 |
| 38 |
245560_at |
AT4G15480
|
UGT84A1 |
0.600 |
0.024 |
| 39 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.581 |
0.064 |
| 40 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.570 |
0.600 |
| 41 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.570 |
0.139 |
| 42 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.569 |
0.057 |
| 43 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.564 |
-0.114 |
| 44 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.563 |
0.945 |
| 45 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.557 |
1.167 |
| 46 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.552 |
1.201 |
| 47 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.550 |
1.168 |
| 48 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.549 |
-0.037 |
| 49 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.549 |
0.112 |
| 50 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.549 |
0.315 |
| 51 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
0.542 |
0.169 |
| 52 |
263829_at |
AT2G40435
|
unknown |
0.541 |
-0.349 |
| 53 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.536 |
1.160 |
| 54 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.535 |
0.279 |
| 55 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.533 |
0.035 |
| 56 |
262452_at |
AT1G11210
|
unknown |
0.533 |
0.376 |
| 57 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.530 |
0.712 |
| 58 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.528 |
0.708 |
| 59 |
253322_at |
AT4G33980
|
unknown |
0.519 |
0.253 |
| 60 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.513 |
0.346 |
| 61 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.512 |
0.055 |
| 62 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.510 |
0.652 |
| 63 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.505 |
0.208 |
| 64 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.504 |
0.146 |
| 65 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.504 |
0.000 |
| 66 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.503 |
0.447 |
| 67 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.500 |
-0.007 |
| 68 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.500 |
0.325 |
| 69 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.499 |
0.021 |
| 70 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.496 |
0.785 |
| 71 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.492 |
0.086 |
| 72 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.492 |
-0.454 |
| 73 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.484 |
-0.101 |
| 74 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.483 |
-0.193 |
| 75 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.476 |
0.083 |
| 76 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.476 |
0.221 |
| 77 |
267036_at |
AT2G38465
|
unknown |
0.475 |
0.200 |
| 78 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.471 |
0.302 |
| 79 |
252661_at |
AT3G44450
|
unknown |
0.468 |
-0.334 |
| 80 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.465 |
-0.303 |
| 81 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.465 |
1.001 |
| 82 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.465 |
0.574 |
| 83 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.464 |
0.298 |
| 84 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.464 |
-0.054 |
| 85 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.461 |
0.743 |
| 86 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.460 |
0.923 |
| 87 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.460 |
0.342 |
| 88 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.459 |
0.211 |
| 89 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.456 |
0.999 |
| 90 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.454 |
0.090 |
| 91 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.454 |
0.544 |
| 92 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.451 |
1.121 |
| 93 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.450 |
-0.194 |
| 94 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.449 |
0.074 |
| 95 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.448 |
-0.091 |
| 96 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.447 |
0.918 |
| 97 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.446 |
0.522 |
| 98 |
260264_at |
AT1G68500
|
unknown |
0.444 |
0.152 |
| 99 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.442 |
0.665 |
| 100 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.442 |
0.209 |
| 101 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.441 |
0.822 |
| 102 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.440 |
0.114 |
| 103 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
0.440 |
0.073 |
| 104 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.439 |
0.092 |
| 105 |
259385_at |
AT1G13470
|
unknown |
0.439 |
0.067 |
| 106 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.438 |
0.065 |
| 107 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.436 |
0.227 |
| 108 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.434 |
0.009 |
| 109 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.434 |
0.231 |
| 110 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.431 |
0.665 |
| 111 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
0.430 |
0.206 |
| 112 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.427 |
0.509 |
| 113 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.427 |
-0.021 |
| 114 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.422 |
0.397 |
| 115 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.421 |
0.318 |
| 116 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.416 |
0.565 |
| 117 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.416 |
-0.010 |
| 118 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
0.413 |
0.223 |
| 119 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.410 |
-0.307 |
| 120 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.409 |
0.858 |
| 121 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.409 |
0.352 |
| 122 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.408 |
-0.225 |
| 123 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.407 |
0.061 |
| 124 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.406 |
0.003 |
| 125 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.405 |
0.123 |
| 126 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.402 |
0.059 |
| 127 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.399 |
-0.097 |
| 128 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.399 |
0.549 |
| 129 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.398 |
0.412 |
| 130 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.396 |
0.687 |
| 131 |
250464_at |
AT5G10040
|
unknown |
-0.755 |
0.048 |
| 132 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.614 |
0.131 |
| 133 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.599 |
0.055 |
| 134 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.476 |
-0.030 |
| 135 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.472 |
0.757 |
| 136 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.450 |
-0.170 |
| 137 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
-0.392 |
0.114 |
| 138 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.391 |
-0.158 |
| 139 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.363 |
-0.272 |
| 140 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.356 |
0.042 |
| 141 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.349 |
0.069 |
| 142 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.347 |
0.100 |
| 143 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.318 |
0.039 |
| 144 |
256710_at |
AT3G30350
|
GLV3, GOLVEN 3, RGF4, root meristem growth factor 4 |
-0.318 |
0.213 |
| 145 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.313 |
0.077 |
| 146 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.312 |
0.091 |
| 147 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.301 |
-0.058 |
| 148 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.288 |
-0.063 |
| 149 |
256926_at |
AT3G22540
|
unknown |
-0.287 |
0.026 |
| 150 |
247024_at |
AT5G66985
|
unknown |
-0.278 |
0.018 |
| 151 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.272 |
0.971 |
| 152 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.270 |
-0.014 |
| 153 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.269 |
0.130 |
| 154 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.267 |
-0.322 |
| 155 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.265 |
0.688 |
| 156 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.265 |
0.064 |
| 157 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.258 |
-0.173 |
| 158 |
259996_at |
AT1G67910
|
unknown |
-0.250 |
0.009 |
| 159 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.250 |
0.059 |
| 160 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.249 |
0.100 |
| 161 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.248 |
-0.368 |
| 162 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.245 |
0.214 |
| 163 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.241 |
0.008 |
| 164 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.240 |
0.375 |
| 165 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.239 |
-0.004 |
| 166 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.238 |
-0.099 |
| 167 |
249206_at |
AT5G42630
|
ATS, ABERRANT TESTA SHAPE, KAN4, KANADI 4 |
-0.235 |
0.057 |
| 168 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.233 |
0.109 |
| 169 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.233 |
-0.137 |
| 170 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
-0.231 |
0.062 |
| 171 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.227 |
0.013 |
| 172 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.227 |
0.012 |
| 173 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
-0.225 |
0.039 |
| 174 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.224 |
-0.051 |
| 175 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.223 |
0.074 |
| 176 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.222 |
-0.119 |
| 177 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.212 |
0.061 |
| 178 |
254145_at |
AT4G24700
|
unknown |
-0.210 |
-0.449 |
| 179 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.204 |
0.043 |
| 180 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
-0.204 |
0.074 |
| 181 |
260472_at |
AT1G10990
|
unknown |
-0.202 |
0.211 |
| 182 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
-0.201 |
0.051 |
| 183 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.200 |
-0.170 |
| 184 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.199 |
0.026 |
| 185 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.199 |
0.053 |
| 186 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.198 |
0.036 |
| 187 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.198 |
0.079 |
| 188 |
256132_at |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.197 |
-0.002 |
| 189 |
263238_at |
AT2G16580
|
SAUR8, SMALL AUXIN UPREGULATED RNA 8 |
-0.196 |
0.045 |
| 190 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.194 |
0.117 |
| 191 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.190 |
0.067 |
| 192 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.190 |
0.033 |
| 193 |
262952_at |
AT1G75770
|
unknown |
-0.189 |
0.054 |
| 194 |
251050_at |
AT5G02440
|
unknown |
-0.186 |
0.060 |
| 195 |
249136_at |
AT5G43180
|
unknown |
-0.186 |
0.197 |
| 196 |
245990_at |
AT5G20640
|
unknown |
-0.184 |
-0.036 |
| 197 |
263769_at |
AT2G06390
|
transposable element gene |
-0.182 |
-0.042 |
| 198 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.181 |
-0.312 |
| 199 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.180 |
0.066 |
| 200 |
256789_at |
AT3G13672
|
SINA2 |
-0.179 |
0.048 |
| 201 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
-0.179 |
0.014 |
| 202 |
263850_at |
AT2G04480
|
unknown |
-0.177 |
0.047 |
| 203 |
267216_at |
AT2G02620
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.176 |
0.112 |
| 204 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.176 |
-0.025 |
| 205 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
-0.176 |
0.214 |
| 206 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.175 |
0.033 |
| 207 |
262421_at |
AT1G50290
|
unknown |
-0.175 |
0.107 |
| 208 |
257946_at |
AT3G21710
|
unknown |
-0.174 |
0.093 |
| 209 |
258468_at |
AT3G06070
|
unknown |
-0.173 |
-0.535 |
| 210 |
251608_at |
AT3G57860
|
GIG1, GIGAS CELL 1, OSD1, OMISSION OF SECOND DIVISION, UVI4-LIKE, UV-B-insensitive 4-like |
-0.172 |
0.032 |
| 211 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.172 |
0.165 |
| 212 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
-0.172 |
-0.165 |
| 213 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.172 |
0.115 |
| 214 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.170 |
0.069 |
| 215 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.169 |
-0.041 |
| 216 |
257551_at |
AT3G13270
|
unknown |
-0.168 |
0.032 |
| 217 |
256262_at |
AT3G12150
|
unknown |
-0.168 |
-0.335 |
| 218 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.167 |
-0.066 |
| 219 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
-0.166 |
0.133 |
| 220 |
257617_at |
AT3G26550
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.166 |
-0.006 |
| 221 |
260341_at |
AT1G69170
|
AtSPL6, SPL6, SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-domain transcription factor 6 |
-0.166 |
-0.032 |
| 222 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
-0.164 |
0.052 |
| 223 |
261345_at |
AT1G79760
|
DTA4, downstream target of AGL15-4 |
-0.161 |
0.153 |
| 224 |
261807_at |
AT1G30515
|
unknown |
-0.161 |
0.067 |
| 225 |
266125_at |
AT2G45050
|
GATA2, GATA transcription factor 2 |
-0.159 |
0.019 |
| 226 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.159 |
0.069 |
| 227 |
258704_at |
AT3G09780
|
ATCRR1, CCR1, CRINKLY4 related 1 |
-0.159 |
0.006 |
| 228 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-0.158 |
-0.026 |
| 229 |
260760_at |
AT1G49170
|
unknown |
-0.158 |
-0.137 |
| 230 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.157 |
-0.048 |
| 231 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.157 |
0.086 |
| 232 |
251412_at |
AT3G60220
|
ATL4, TOXICOS EN LEVADURA 4, TL4, TOXICOS EN LEVADURA 4 |
-0.157 |
0.009 |
| 233 |
248674_at |
AT5G48800
|
[Phototropic-responsive NPH3 family protein] |
-0.157 |
-0.120 |
| 234 |
253394_at |
AT4G32770
|
ATSDX1, SUCROSE EXPORT DEFECTIVE 1, VTE1, VITAMIN E DEFICIENT 1 |
-0.156 |
-0.081 |
| 235 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.156 |
-0.186 |
| 236 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
-0.156 |
0.126 |
| 237 |
248535_at |
AT5G50120
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.155 |
-0.041 |
| 238 |
262388_at |
AT1G49320
|
unknown |
-0.154 |
-0.068 |
| 239 |
249389_at |
AT5G40100
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.153 |
0.293 |
| 240 |
258424_at |
AT3G16750
|
unknown |
-0.150 |
-0.210 |
| 241 |
264586_at |
AT1G05160
|
ATKAO1, CYP88A3, cytochrome P450, family 88, subfamily A, polypeptide 3, KAO1, ENT-KAURENOIC ACID OXYDASE 1 |
-0.150 |
0.130 |
| 242 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.149 |
-0.154 |
| 243 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.149 |
-0.068 |
| 244 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.148 |
-0.137 |
| 245 |
247570_at |
AT5G61250
|
AtGUS1, glucuronidase 1, GUS1, glucuronidase 1 |
-0.148 |
0.129 |
| 246 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
-0.148 |
0.042 |
| 247 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.147 |
-0.243 |
| 248 |
262121_at |
AT1G02800
|
ATCEL2, cellulase 2, CEL2, CEL2, cellulase 2 |
-0.147 |
0.131 |
| 249 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.146 |
0.151 |
| 250 |
245513_at |
AT4G15780
|
ATVAMP724, vesicle-associated membrane protein 724, VAMP724, vesicle-associated membrane protein 724 |
-0.146 |
0.177 |
| 251 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.145 |
-0.020 |
| 252 |
265683_at |
AT2G24400
|
SAUR38, SMALL AUXIN UPREGULATED RNA 38 |
-0.145 |
0.028 |
| 253 |
267077_at |
AT2G40970
|
MYBC1 |
-0.145 |
-0.223 |
| 254 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
-0.145 |
-0.070 |
| 255 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.143 |
-0.330 |
| 256 |
246063_at |
AT5G19340
|
unknown |
-0.142 |
0.082 |
| 257 |
246244_at |
AT4G37250
|
[Leucine-rich repeat protein kinase family protein] |
-0.140 |
0.015 |
| 258 |
250481_at |
AT5G10310
|
unknown |
-0.139 |
0.064 |
| 259 |
264836_at |
AT1G03610
|
unknown |
-0.139 |
0.133 |