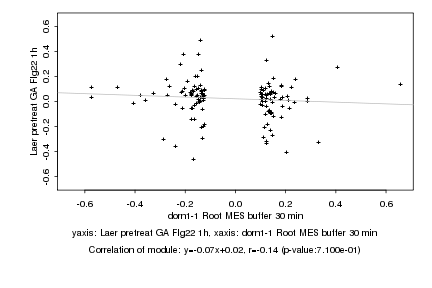
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
dorn1-1 Root MES buffer 30 min |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
252545_at |
AT3G45820
|
unknown |
0.656 |
0.138 |
| 2 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.406 |
0.275 |
| 3 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.327 |
-0.320 |
| 4 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.287 |
0.025 |
| 5 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.284 |
0.006 |
| 6 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
0.239 |
0.182 |
| 7 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.235 |
-0.002 |
| 8 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.221 |
0.118 |
| 9 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.212 |
-0.053 |
| 10 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.210 |
0.011 |
| 11 |
267636_at |
AT2G42110
|
unknown |
0.207 |
0.042 |
| 12 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.203 |
-0.403 |
| 13 |
264763_at |
AT1G61450
|
unknown |
0.187 |
0.040 |
| 14 |
252261_at |
AT3G49500
|
RDR6, RNA-dependent RNA polymerase 6, SDE1, SILENCING DEFECTIVE 1, SGS2, SUPPRESSOR OF GENE SILENCING 2 |
0.186 |
-0.037 |
| 15 |
253055_at |
AT4G37630
|
CYCD5;1, cyclin d5;1 |
0.182 |
0.125 |
| 16 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
0.181 |
-0.122 |
| 17 |
263474_at |
AT2G31725
|
unknown |
0.181 |
0.136 |
| 18 |
254391_at |
AT4G21590
|
ENDO3, endonuclease 3 |
0.178 |
0.017 |
| 19 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.157 |
0.065 |
| 20 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.152 |
0.037 |
| 21 |
250198_at |
AT5G14100
|
ABCI11, ATP-binding cassette I11, ATNAP14, ARABIDOPSIS THALIANANON-INTRINSIC ABC PROTEIN 14, NAP14, non-intrinsic ABC protein 14 |
0.150 |
-0.119 |
| 22 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.149 |
0.185 |
| 23 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.149 |
0.074 |
| 24 |
260683_at |
AT1G17560
|
HLL, HUELLENLOS |
0.145 |
-0.005 |
| 25 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.145 |
-0.265 |
| 26 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.145 |
0.521 |
| 27 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.143 |
0.087 |
| 28 |
255583_at |
AT4G01510
|
ARV2 |
0.142 |
-0.093 |
| 29 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.140 |
-0.084 |
| 30 |
255887_at |
AT1G20370
|
[Pseudouridine synthase family protein] |
0.139 |
-0.094 |
| 31 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.138 |
0.067 |
| 32 |
254181_at |
AT4G23940
|
ARC1, ACCUMULATION AND REPLICATION OF CHLOROPLASTS 1, FtsHi1, FTSH inactive protease 1 |
0.138 |
-0.230 |
| 33 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
0.137 |
0.023 |
| 34 |
251576_at |
AT3G58200
|
[TRAF-like family protein] |
0.136 |
0.060 |
| 35 |
252946_at |
AT4G39235
|
unknown |
0.135 |
-0.077 |
| 36 |
254182_at |
AT4G23950
|
[Galactose-binding protein] |
0.134 |
0.068 |
| 37 |
248476_at |
AT5G50890
|
[alpha/beta-Hydrolases superfamily protein] |
0.134 |
-0.065 |
| 38 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.133 |
0.123 |
| 39 |
257724_at |
AT3G18510
|
unknown |
0.130 |
-0.076 |
| 40 |
265065_at |
AT1G03980
|
ATPCS2, phytochelatin synthase 2, PCS2, phytochelatin synthase 2 |
0.128 |
0.148 |
| 41 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.127 |
-0.182 |
| 42 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
0.124 |
0.058 |
| 43 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.123 |
-0.314 |
| 44 |
261025_at |
AT1G01225
|
[NC domain-containing protein-related] |
0.122 |
0.050 |
| 45 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
0.122 |
-0.033 |
| 46 |
246808_at |
AT5G27110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.121 |
0.026 |
| 47 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.120 |
-0.332 |
| 48 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.120 |
0.335 |
| 49 |
246731_at |
AT5G27630
|
ACBP5, acyl-CoA binding protein 5, AtACBP5 |
0.119 |
0.107 |
| 50 |
255558_at |
AT4G01900
|
GLB1, GLNB1 homolog, PII |
0.119 |
-0.101 |
| 51 |
253726_at |
AT4G29430
|
rps15ae, ribosomal protein S15A E |
0.119 |
0.058 |
| 52 |
265646_at |
AT2G27360
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.117 |
0.008 |
| 53 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
0.114 |
-0.201 |
| 54 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
0.108 |
0.090 |
| 55 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.108 |
-0.285 |
| 56 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
0.106 |
-0.027 |
| 57 |
253782_at |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
0.106 |
0.037 |
| 58 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
0.105 |
0.033 |
| 59 |
251574_at |
AT3G58100
|
PDCB5, plasmodesmata callose-binding protein 5 |
0.104 |
0.059 |
| 60 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
0.104 |
0.001 |
| 61 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
0.103 |
0.041 |
| 62 |
253085_s_at |
AT4G36280
|
CRH1, CRT1 Homologue 1, MORC2, microrchidia 2 |
0.102 |
0.061 |
| 63 |
263630_at |
AT2G04845
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.102 |
0.099 |
| 64 |
266264_at |
AT2G27775
|
unknown |
0.102 |
0.116 |
| 65 |
246269_at |
AT4G37110
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
0.101 |
0.061 |
| 66 |
256484_at |
AT1G31430
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.100 |
0.017 |
| 67 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
0.100 |
0.023 |
| 68 |
249060_at |
AT5G44560
|
VPS2.2 |
0.099 |
-0.017 |
| 69 |
265824_at |
AT2G35650
|
ATCSLA07, cellulose synthase like, ATCSLA7, CSLA07, CSLA07, cellulose synthase like, CSLA7, CELLULOSE SYNTHASE LIKE A7 |
0.099 |
0.073 |
| 70 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.576 |
0.036 |
| 71 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.575 |
0.120 |
| 72 |
260495_at |
AT2G41810
|
unknown |
-0.473 |
0.117 |
| 73 |
266900_at |
AT2G34610
|
unknown |
-0.410 |
-0.015 |
| 74 |
267228_at |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
-0.382 |
0.056 |
| 75 |
256162_at |
AT1G55390
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.359 |
0.011 |
| 76 |
261675_at |
AT1G18290
|
unknown |
-0.329 |
0.069 |
| 77 |
262388_at |
AT1G49320
|
unknown |
-0.289 |
-0.298 |
| 78 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.275 |
0.184 |
| 79 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.271 |
0.051 |
| 80 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.266 |
0.128 |
| 81 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.241 |
-0.352 |
| 82 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.240 |
-0.017 |
| 83 |
264680_at |
AT1G65510
|
unknown |
-0.220 |
0.300 |
| 84 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
-0.215 |
0.073 |
| 85 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.214 |
0.082 |
| 86 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.211 |
-0.048 |
| 87 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
-0.207 |
0.384 |
| 88 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
-0.204 |
0.109 |
| 89 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
-0.199 |
0.051 |
| 90 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.193 |
0.164 |
| 91 |
265685_at |
AT2G24430
|
ANAC038, NAC domain containing protein 38, ANAC039, Arabidopsis NAC domain containing protein 39, NAC038, NAC domain containing protein 38 |
-0.182 |
0.075 |
| 92 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.179 |
-0.053 |
| 93 |
249562_at |
AT5G38350
|
[Disease resistance protein (NBS-LRR class) family] |
-0.179 |
0.064 |
| 94 |
267343_at |
AT2G44260
|
unknown |
-0.178 |
0.049 |
| 95 |
263282_at |
AT2G14095
|
unknown |
-0.177 |
0.069 |
| 96 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.176 |
-0.139 |
| 97 |
259586_at |
AT1G28100
|
unknown |
-0.174 |
-0.050 |
| 98 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
-0.172 |
0.089 |
| 99 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.168 |
-0.460 |
| 100 |
257458_at |
AT2G05400
|
[Ubiquitin-specific protease family C19-related protein] |
-0.168 |
0.081 |
| 101 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-0.167 |
-0.138 |
| 102 |
257026_at |
AT3G19200
|
unknown |
-0.165 |
0.127 |
| 103 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.164 |
-0.028 |
| 104 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.162 |
0.203 |
| 105 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
-0.158 |
0.047 |
| 106 |
252286_at |
AT3G49070
|
unknown |
-0.158 |
0.103 |
| 107 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.158 |
-0.010 |
| 108 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
-0.155 |
0.204 |
| 109 |
261393_at |
AT1G79580
|
ANAC033, Arabidopsis NAC domain containing protein 33, SMB, SOMBRERO, URP7, UAS-TAGGED ROOT PATTERNING7 |
-0.155 |
0.056 |
| 110 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.150 |
0.023 |
| 111 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.149 |
0.110 |
| 112 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.148 |
0.377 |
| 113 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.146 |
0.006 |
| 114 |
260058_at |
AT1G78100
|
AUF1, auxin up-regulated f-box protein 1 |
-0.145 |
-0.001 |
| 115 |
253662_at |
AT4G30080
|
ARF16, auxin response factor 16 |
-0.142 |
0.039 |
| 116 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
-0.140 |
0.131 |
| 117 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.140 |
0.494 |
| 118 |
265546_at |
AT2G28240
|
[ATP-dependent helicase family protein] |
-0.140 |
0.004 |
| 119 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.138 |
0.094 |
| 120 |
256515_at |
AT1G66020
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.137 |
0.066 |
| 121 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
-0.136 |
0.254 |
| 122 |
248140_at |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
-0.136 |
-0.207 |
| 123 |
247686_at |
AT5G59700
|
[Protein kinase superfamily protein] |
-0.136 |
0.081 |
| 124 |
250810_at |
AT5G05090
|
[Homeodomain-like superfamily protein] |
-0.135 |
0.066 |
| 125 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.133 |
-0.060 |
| 126 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.132 |
-0.292 |
| 127 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
-0.132 |
0.070 |
| 128 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.131 |
0.027 |
| 129 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
-0.131 |
-0.193 |
| 130 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.130 |
0.031 |
| 131 |
250223_at |
AT5G14070
|
ROXY2 |
-0.129 |
0.046 |
| 132 |
266844_at |
AT2G26120
|
[glycine-rich protein] |
-0.128 |
0.015 |
| 133 |
254561_at |
AT4G19160
|
unknown |
-0.128 |
0.056 |
| 134 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.127 |
0.095 |
| 135 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.126 |
0.055 |
| 136 |
251188_at |
AT3G62730
|
unknown |
-0.126 |
0.101 |
| 137 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.126 |
-0.181 |