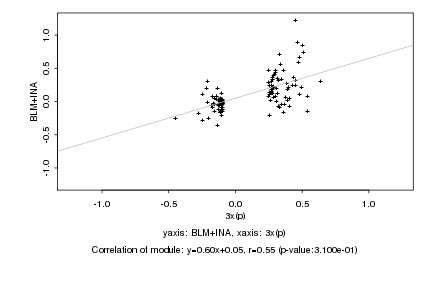
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
3x(p) |
Log2 signal ratio
BLM+INA |
| 1 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.637 |
0.307 |
| 2 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.539 |
-0.138 |
| 3 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.537 |
0.082 |
| 4 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.504 |
0.749 |
| 5 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.501 |
0.844 |
| 6 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.491 |
0.222 |
| 7 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.477 |
0.668 |
| 8 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.473 |
0.107 |
| 9 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.468 |
0.596 |
| 10 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.462 |
0.893 |
| 11 |
264774_at |
AT1G22890
|
unknown |
0.447 |
0.330 |
| 12 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.443 |
0.246 |
| 13 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.443 |
1.234 |
| 14 |
265387_at |
AT2G20670
|
unknown |
0.432 |
0.370 |
| 15 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.425 |
0.248 |
| 16 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.405 |
-0.069 |
| 17 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.400 |
0.047 |
| 18 |
254954_at |
AT4G10910
|
unknown |
0.395 |
0.224 |
| 19 |
266259_at |
AT2G27830
|
unknown |
0.388 |
0.185 |
| 20 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.385 |
0.018 |
| 21 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.376 |
0.282 |
| 22 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.375 |
0.066 |
| 23 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.364 |
-0.033 |
| 24 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.355 |
0.470 |
| 25 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.355 |
-0.155 |
| 26 |
263182_at |
AT1G05575
|
unknown |
0.339 |
0.343 |
| 27 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.339 |
-0.043 |
| 28 |
266658_at |
AT2G25735
|
unknown |
0.337 |
0.568 |
| 29 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.326 |
-0.085 |
| 30 |
256442_at |
AT3G10930
|
unknown |
0.323 |
0.715 |
| 31 |
265184_at |
AT1G23710
|
unknown |
0.320 |
0.317 |
| 32 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.318 |
-0.065 |
| 33 |
245041_at |
AT2G26530
|
AR781 |
0.313 |
0.359 |
| 34 |
258939_at |
AT3G10020
|
unknown |
0.311 |
0.127 |
| 35 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
0.302 |
0.001 |
| 36 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.302 |
0.211 |
| 37 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.297 |
0.449 |
| 38 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.297 |
0.089 |
| 39 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.293 |
0.470 |
| 40 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.288 |
0.421 |
| 41 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.285 |
0.229 |
| 42 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.284 |
0.218 |
| 43 |
264379_at |
AT2G25200
|
unknown |
0.280 |
0.402 |
| 44 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.280 |
0.068 |
| 45 |
259015_at |
AT3G07350
|
unknown |
0.277 |
0.183 |
| 46 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.277 |
0.367 |
| 47 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.274 |
0.337 |
| 48 |
254178_at |
AT4G23880
|
unknown |
0.272 |
0.338 |
| 49 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.271 |
0.124 |
| 50 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.270 |
0.193 |
| 51 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.269 |
0.143 |
| 52 |
259479_at |
AT1G19020
|
unknown |
0.269 |
0.251 |
| 53 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.269 |
0.215 |
| 54 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.268 |
0.153 |
| 55 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.265 |
0.133 |
| 56 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.265 |
0.312 |
| 57 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.262 |
0.018 |
| 58 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
0.252 |
-0.209 |
| 59 |
258262_at |
AT3G15770
|
unknown |
0.250 |
0.111 |
| 60 |
248432_at |
AT5G51390
|
unknown |
0.249 |
0.252 |
| 61 |
247585_at |
AT5G60680
|
unknown |
0.248 |
0.145 |
| 62 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.245 |
0.477 |
| 63 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.245 |
0.084 |
| 64 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.243 |
0.290 |
| 65 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.454 |
-0.244 |
| 66 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.283 |
-0.174 |
| 67 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.252 |
0.113 |
| 68 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.249 |
-0.284 |
| 69 |
254571_at |
AT4G19370
|
unknown |
-0.220 |
0.206 |
| 70 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
-0.215 |
-0.011 |
| 71 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
-0.212 |
0.304 |
| 72 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.207 |
-0.250 |
| 73 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.182 |
-0.051 |
| 74 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.179 |
0.077 |
| 75 |
255858_at |
AT1G67030
|
ZFP6, zinc finger protein 6 |
-0.177 |
-0.086 |
| 76 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
-0.169 |
-0.023 |
| 77 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.164 |
0.055 |
| 78 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
-0.163 |
-0.034 |
| 79 |
255296_at |
AT4G04790
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.160 |
0.053 |
| 80 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.159 |
-0.136 |
| 81 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.149 |
0.041 |
| 82 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
-0.149 |
0.081 |
| 83 |
252661_at |
AT3G44450
|
unknown |
-0.144 |
0.076 |
| 84 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
-0.137 |
0.199 |
| 85 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.137 |
-0.359 |
| 86 |
259003_at |
AT3G02010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.133 |
-0.057 |
| 87 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
-0.129 |
0.042 |
| 88 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.128 |
-0.038 |
| 89 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
-0.128 |
-0.112 |
| 90 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
-0.125 |
0.012 |
| 91 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.124 |
-0.057 |
| 92 |
253938_at |
AT4G26920
|
[START (StAR-related lipid-transfer) lipid-binding domain] |
-0.121 |
-0.018 |
| 93 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
-0.120 |
0.018 |
| 94 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.120 |
-0.020 |
| 95 |
261750_at |
AT1G76120
|
[Pseudouridine synthase family protein] |
-0.120 |
0.034 |
| 96 |
253247_at |
AT4G34610
|
BLH6, BEL1-like homeodomain 6 |
-0.118 |
-0.143 |
| 97 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.118 |
-0.158 |
| 98 |
253165_at |
AT4G35320
|
unknown |
-0.117 |
-0.161 |
| 99 |
250283_at |
AT5G13270
|
RARE1, REQUIRED FOR ACCD RNA EDITING 1 |
-0.116 |
0.020 |
| 100 |
245584_at |
AT4G14940
|
AO1, amine oxidase 1, ATAO1, amine oxidase 1 |
-0.116 |
-0.162 |
| 101 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.115 |
0.055 |
| 102 |
248857_at |
AT5G46640
|
AHL8, AT-hook motif nuclear localized protein 8 |
-0.114 |
-0.023 |
| 103 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.114 |
-0.154 |
| 104 |
258861_at |
AT3G02060
|
[DEAD/DEAH box helicase, putative] |
-0.113 |
-0.003 |
| 105 |
257852_at |
AT3G12950
|
[Trypsin family protein] |
-0.111 |
0.042 |
| 106 |
262941_at |
AT1G79490
|
EMB2217, embryo defective 2217 |
-0.110 |
0.028 |
| 107 |
266879_at |
AT2G44590
|
ADL1D, DYNAMIN-like 1D, DL1D, DYNAMIN-like 1D |
-0.109 |
0.033 |
| 108 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.109 |
-0.126 |
| 109 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.109 |
-0.075 |
| 110 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.108 |
-0.114 |
| 111 |
254632_at |
AT4G18630
|
unknown |
-0.108 |
-0.198 |
| 112 |
256919_at |
AT3G18970
|
MEF20, mitochondrial editing factor 20 |
-0.107 |
-0.039 |
| 113 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.107 |
0.135 |
| 114 |
252438_at |
AT3G47390
|
PHS1, PHOTOSENSITIVE 1, PyrR, pyrimidine reductase |
-0.105 |
-0.050 |
| 115 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.105 |
-0.031 |
| 116 |
263805_at |
AT2G40400
|
unknown |
-0.104 |
-0.025 |
| 117 |
264816_at |
AT1G03560
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.104 |
0.038 |
| 118 |
259605_at |
AT1G27910
|
ATPUB45, ARABIDOPSIS THALIANA PLANT U-BOX 45, PUB45, plant U-box 45 |
-0.104 |
-0.109 |
| 119 |
253659_at |
AT4G30150
|
unknown |
-0.104 |
-0.003 |
| 120 |
251462_at |
AT3G59790
|
ATMPK10, MAP kinase 10, MPK10, MAP kinase 10 |
-0.103 |
-0.043 |
| 121 |
246580_at |
AT1G31770
|
ABCG14, ATP-binding cassette G14 |
-0.103 |
-0.026 |
| 122 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.103 |
-0.017 |
| 123 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.103 |
-0.079 |
| 124 |
264356_at |
AT1G03190
|
ATXPD, ARABIDOPSIS THALIANA XERODERMA PIGMENTOSUM GROUP D, UVH6, ULTRAVIOLET HYPERSENSITIVE 6 |
-0.102 |
0.024 |
| 125 |
245109_at |
AT2G41520
|
DJC62, DNA J protein C62, TPR15, tetratricopeptide repeat 15 |
-0.102 |
0.014 |
| 126 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.101 |
-0.147 |
| 127 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.099 |
-0.009 |