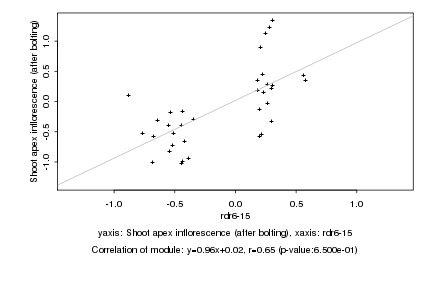
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rdr6-15 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.575 |
0.357 |
| 2 |
246980_at |
AT5G67530
|
plant U-box 49 |
0.554 |
0.440 |
| 3 |
254400_at |
AT4G21270
|
ATK1, kinesin 1, KATA, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A, KATAP, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A PROTEIN |
0.303 |
0.267 |
| 4 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
0.298 |
1.357 |
| 5 |
257145_at |
AT3G27320
|
[alpha/beta-Hydrolases superfamily protein] |
0.292 |
0.220 |
| 6 |
260701_at |
AT1G32330
|
ATHSFA1D, heat shock transcription factor A1D, HSFA1D, heat shock transcription factor A1D |
0.292 |
-0.329 |
| 7 |
267452_at |
AT2G33860
|
ARF3, AUXIN RESPONSE TRANSCRIPTION FACTOR 3, ETT, ETTIN |
0.277 |
1.229 |
| 8 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
0.263 |
0.286 |
| 9 |
255739_at |
AT1G25580
|
ANAC008, Arabidopsis NAC domain containing protein 8, SOG1, SUPPRESSOR OF GAMMA RADIATION 1 |
0.256 |
-0.019 |
| 10 |
250779_at |
AT5G05470
|
ATEIF2-A2, EIF2 ALPHA, eukaryotic translation initiation factor 2 alpha subunit, EIF2-A2 |
0.244 |
1.142 |
| 11 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
0.225 |
0.160 |
| 12 |
253710_at |
AT4G29230
|
anac075, NAC domain containing protein 75, NAC075, NAC domain containing protein 75 |
0.216 |
0.450 |
| 13 |
253426_at |
AT4G32375
|
[Pectin lyase-like superfamily protein] |
0.214 |
-0.532 |
| 14 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
0.205 |
0.899 |
| 15 |
261121_at |
AT1G75340
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.195 |
-0.132 |
| 16 |
262093_at |
AT1G56145
|
[Leucine-rich repeat transmembrane protein kinase] |
0.191 |
-0.564 |
| 17 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.176 |
0.198 |
| 18 |
263730_at |
AT1G60090
|
BGLU4, beta glucosidase 4 |
0.174 |
0.354 |
| 19 |
246967_at |
AT5G24860
|
ATFPF1, ARABIDOPSIS FLOWERING PROMOTING FACTOR 1, FPF1, FLOWERING PROMOTING FACTOR 1 |
-0.883 |
0.116 |
| 20 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
-0.774 |
-0.518 |
| 21 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.685 |
-1.006 |
| 22 |
249454_at |
AT5G39520
|
unknown |
-0.682 |
-0.566 |
| 23 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.645 |
-0.309 |
| 24 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
-0.559 |
-0.390 |
| 25 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.548 |
-0.822 |
| 26 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
-0.540 |
-0.172 |
| 27 |
247547_at |
AT5G61360
|
unknown |
-0.519 |
-0.721 |
| 28 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-0.511 |
-0.521 |
| 29 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.451 |
-0.385 |
| 30 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
-0.450 |
-1.017 |
| 31 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
-0.438 |
-0.150 |
| 32 |
259451_at |
AT1G13890
|
ATSNAP30, SNAP30, soluble N-ethylmaleimide-sensitive factor adaptor protein 30 |
-0.438 |
-0.989 |
| 33 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
-0.420 |
-0.648 |
| 34 |
251433_at |
AT3G59830
|
[Integrin-linked protein kinase family] |
-0.395 |
-0.931 |
| 35 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.352 |
-0.287 |