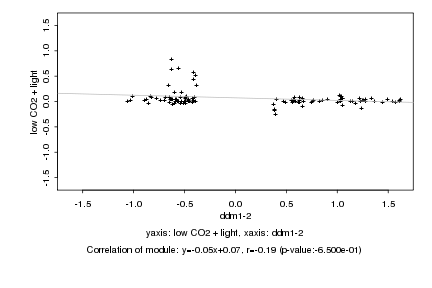
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ddm1-2 |
Log2 signal ratio
low CO2 + light |
| 1 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.617 |
0.045 |
| 2 |
263675_x_at |
AT2G04770
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.0e-20 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.610 |
0.022 |
| 3 |
261619_x_at |
AT1G33130
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.3e-125 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.605 |
0.012 |
| 4 |
255246_at |
AT4G05640
|
unknown |
1.569 |
-0.011 |
| 5 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.541 |
0.018 |
| 6 |
261186_at |
AT1G34530
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.8e-116 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.492 |
0.051 |
| 7 |
245203_at |
AT3G33080
|
unknown |
1.435 |
0.000 |
| 8 |
262268_at |
AT1G42410
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.0e-124 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.365 |
0.008 |
| 9 |
263293_x_at |
AT2G10880
|
[hypothetical protein, similar to hypothetical protein GB:AAC26673] |
1.335 |
0.077 |
| 10 |
256748_x_at |
AT3G30396
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.1e-39 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.277 |
0.054 |
| 11 |
257110_x_at |
AT3G30440
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
1.268 |
0.011 |
| 12 |
255313_at |
AT4G04590
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 5.3e-36 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
1.252 |
0.038 |
| 13 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.241 |
0.024 |
| 14 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
1.235 |
-0.130 |
| 15 |
263479_x_at |
AT2G04000
|
unknown |
1.227 |
0.005 |
| 16 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
1.218 |
0.071 |
| 17 |
249259_at |
AT5G41660
|
unknown |
1.173 |
-0.023 |
| 18 |
258302_at |
AT3G30610
|
unknown |
1.143 |
0.012 |
| 19 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
1.125 |
0.010 |
| 20 |
245032_at |
AT2G26630
|
[transposase IS4 family protein, contains Pfam profile: PF01609 transposase DDE domain] |
1.050 |
-0.061 |
| 21 |
257777_x_at |
AT3G29210
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
1.049 |
0.062 |
| 22 |
257111_x_at |
AT3G30450
|
[similar to heat shock protein binding [Arabidopsis thaliana] (TAIR:AT3G30450.1)] |
1.042 |
0.061 |
| 23 |
246640_x_at |
AT5G34900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
1.038 |
0.104 |
| 24 |
263307_at |
AT2G12520
|
unknown |
1.025 |
0.004 |
| 25 |
246639_x_at |
AT5G34895
|
[similar to heat shock protein binding [Arabidopsis thaliana] (TAIR:AT1G32830.1)] |
1.025 |
0.117 |
| 26 |
250979_at |
AT5G03090
|
unknown |
1.023 |
0.050 |
| 27 |
261235_x_at |
AT1G32840
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
1.014 |
0.125 |
| 28 |
259572_at |
AT1G20400
|
unknown |
1.001 |
-0.016 |
| 29 |
263346_at |
AT2G05660
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.898 |
0.059 |
| 30 |
249030_at |
AT5G44880
|
unknown |
0.849 |
0.034 |
| 31 |
264995_at |
AT1G67240
|
[Mutator-like transposase family, has a 4.5e-23 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.820 |
0.019 |
| 32 |
255917_at |
AT5G28560
|
unknown |
0.757 |
0.022 |
| 33 |
256449_at |
AT1G33460
|
[Mutator-like transposase family, has a 3.6e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.755 |
0.004 |
| 34 |
248949_at |
AT5G45570
|
[Ulp1 protease family protein] |
0.746 |
-0.007 |
| 35 |
249114_at |
AT5G43800
|
[copia-like retrotransposon family, has a 0. P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.665 |
0.020 |
| 36 |
255932_at |
AT1G12720
|
[Mutator-like transposase family, has a 1.2e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.657 |
0.060 |
| 37 |
258370_at |
AT3G14395
|
unknown |
0.651 |
-0.094 |
| 38 |
260188_at |
AT1G35995
|
[Mutator-like transposase family, has a 4.4e-38 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.626 |
-0.000 |
| 39 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.625 |
0.037 |
| 40 |
257354_x_at |
AT2G23480
|
unknown |
0.621 |
0.095 |
| 41 |
255120_x_at |
AT4G08890
|
[Mutator-like transposase family, has a 5.6e-82 P-value blast match to O65231 /281-442 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.614 |
-0.006 |
| 42 |
262256_at |
AT1G53810
|
[copia-like retrotransposon family, has a 8.0e-248 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
0.581 |
0.006 |
| 43 |
249737_at |
AT5G24480
|
[Beta-galactosidase related protein] |
0.577 |
0.081 |
| 44 |
262034_at |
AT1G37080
|
[similar to myb family protein [Arabidopsis thaliana] (TAIR:AT2G22710.1)] |
0.564 |
0.022 |
| 45 |
263817_at |
AT2G10070
|
unknown |
0.561 |
0.049 |
| 46 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
0.559 |
-0.006 |
| 47 |
254671_at |
AT4G18410
|
[Mutator-like transposase family, has a 8.7e-30 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.549 |
0.038 |
| 48 |
255476_at |
AT4G02490
|
[non-LTR retrotransposon family (LINE), has a 2.3e-36 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
0.483 |
-0.013 |
| 49 |
263273_x_at |
AT2G11590
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.468 |
0.018 |
| 50 |
267440_at |
AT2G19070
|
SHT, spermidine hydroxycinnamoyl transferase |
0.402 |
0.046 |
| 51 |
258716_at |
AT3G09700
|
[Chaperone DnaJ-domain superfamily protein] |
0.392 |
-0.255 |
| 52 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.379 |
-0.147 |
| 53 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
0.374 |
-0.170 |
| 54 |
261285_at |
AT1G35720
|
ANN1, annexin 1, ANNAT1, annexin 1, AtANN1, ATOXY5, OXY5 |
0.366 |
-0.052 |
| 55 |
257469_at |
AT1G49290
|
unknown |
-1.067 |
0.014 |
| 56 |
262282_at |
AT1G68610
|
PCR11, PLANT CADMIUM RESISTANCE 11 |
-1.037 |
0.022 |
| 57 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
-1.013 |
0.110 |
| 58 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
-0.895 |
0.034 |
| 59 |
259464_at |
AT1G18990
|
unknown |
-0.874 |
0.046 |
| 60 |
257119_at |
AT3G20190
|
PRK4, pollen receptor like kinase 4 |
-0.861 |
-0.028 |
| 61 |
251697_at |
AT3G56600
|
[Protein kinase superfamily protein] |
-0.839 |
0.101 |
| 62 |
246123_at |
AT5G20390
|
[Glycosyl hydrolase superfamily protein] |
-0.833 |
0.083 |
| 63 |
257804_at |
AT3G18810
|
AtPERK6, proline-rich extensin-like receptor kinase 6, PERK6, proline-rich extensin-like receptor kinase 6 |
-0.779 |
0.070 |
| 64 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
-0.737 |
0.039 |
| 65 |
259886_at |
AT1G76370
|
[Protein kinase superfamily protein] |
-0.699 |
0.024 |
| 66 |
245232_at |
AT4G25590
|
ADF7, actin depolymerizing factor 7 |
-0.693 |
0.092 |
| 67 |
246789_at |
AT5G27600
|
ATLACS7, LACS7, long-chain acyl-CoA synthetase 7 |
-0.662 |
0.328 |
| 68 |
250121_at |
AT5G16500
|
LIP1, LOST IN POLLEN TUBE GUIDANCE 1 |
-0.653 |
0.000 |
| 69 |
245057_at |
AT2G26490
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.653 |
0.080 |
| 70 |
250466_at |
AT5G10090
|
TPR13, tetratricopeptide repeat 13 |
-0.640 |
0.052 |
| 71 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.632 |
0.644 |
| 72 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.630 |
0.838 |
| 73 |
255835_at |
AT2G33420
|
unknown |
-0.629 |
0.057 |
| 74 |
266453_at |
AT2G43230
|
[Protein kinase superfamily protein] |
-0.623 |
0.058 |
| 75 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
-0.621 |
-0.046 |
| 76 |
256021_at |
AT1G58270
|
ZW9 |
-0.605 |
0.196 |
| 77 |
262620_at |
AT1G06540
|
unknown |
-0.601 |
-0.037 |
| 78 |
263062_at |
AT2G18180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.597 |
0.059 |
| 79 |
254829_at |
AT4G12530
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.590 |
0.007 |
| 80 |
256506_at |
AT1G75160
|
unknown |
-0.581 |
0.056 |
| 81 |
260737_at |
AT1G17540
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.576 |
0.019 |
| 82 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
-0.566 |
0.017 |
| 83 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.562 |
0.655 |
| 84 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.543 |
0.085 |
| 85 |
254762_at |
AT4G13230
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.543 |
-0.031 |
| 86 |
247547_at |
AT5G61360
|
unknown |
-0.539 |
0.002 |
| 87 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.539 |
0.181 |
| 88 |
249194_at |
AT5G42490
|
[ATP binding microtubule motor family protein] |
-0.514 |
-0.027 |
| 89 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
-0.505 |
0.007 |
| 90 |
260193_at |
AT1G67640
|
[Transmembrane amino acid transporter family protein] |
-0.497 |
0.068 |
| 91 |
264139_at |
AT1G78940
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.493 |
-0.028 |
| 92 |
265681_at |
AT2G24370
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.493 |
0.002 |
| 93 |
267438_at |
AT2G19050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.492 |
0.093 |
| 94 |
267322_at |
AT2G19330
|
PIRL6, plant intracellular ras group-related LRR 6 |
-0.486 |
0.110 |
| 95 |
252531_at |
AT3G46520
|
ACT12, actin-12 |
-0.470 |
0.011 |
| 96 |
259338_at |
AT3G03800
|
ATSYP131, SYP131, syntaxin of plants 131 |
-0.467 |
0.027 |
| 97 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.464 |
0.048 |
| 98 |
259451_at |
AT1G13890
|
ATSNAP30, SNAP30, soluble N-ethylmaleimide-sensitive factor adaptor protein 30 |
-0.454 |
0.014 |
| 99 |
256261_at |
AT3G12160
|
ATRABA4D, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A4D, RABA4D, RAB GTPase homolog A4D |
-0.427 |
-0.017 |
| 100 |
256782_at |
AT3G13660
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.423 |
0.065 |
| 101 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.420 |
0.592 |
| 102 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
-0.418 |
0.453 |
| 103 |
251293_at |
AT3G61930
|
unknown |
-0.412 |
0.028 |
| 104 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.403 |
0.099 |
| 105 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.400 |
0.515 |
| 106 |
251711_at |
AT3G56960
|
PIP5K4, phosphatidyl inositol monophosphate 5 kinase 4 |
-0.397 |
0.016 |
| 107 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.392 |
0.319 |