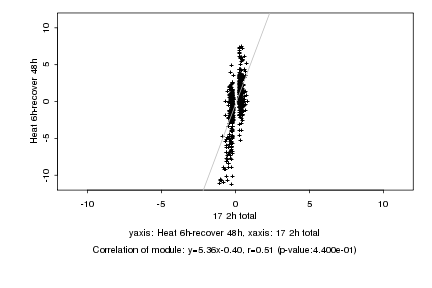
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
17 2h total |
Log2 signal ratio
Heat 6h-recover 48h |
| 1 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.751 |
0.121 |
| 2 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.696 |
0.822 |
| 3 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.683 |
5.186 |
| 4 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.637 |
4.197 |
| 5 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.635 |
1.468 |
| 6 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.632 |
-1.112 |
| 7 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.615 |
-0.343 |
| 8 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.612 |
3.600 |
| 9 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
0.607 |
6.148 |
| 10 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.604 |
1.317 |
| 11 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.600 |
1.348 |
| 12 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.597 |
-0.415 |
| 13 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.596 |
4.458 |
| 14 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.580 |
0.288 |
| 15 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.576 |
0.121 |
| 16 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.553 |
3.367 |
| 17 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.546 |
0.706 |
| 18 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.542 |
2.222 |
| 19 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.529 |
3.331 |
| 20 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.518 |
0.372 |
| 21 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
0.510 |
0.121 |
| 22 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
0.498 |
0.121 |
| 23 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.492 |
-1.317 |
| 24 |
261247_at |
AT1G20070
|
unknown |
0.490 |
2.409 |
| 25 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.489 |
-0.040 |
| 26 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
0.487 |
3.444 |
| 27 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.483 |
-0.637 |
| 28 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.478 |
3.549 |
| 29 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
0.472 |
1.213 |
| 30 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.469 |
1.788 |
| 31 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
0.461 |
7.241 |
| 32 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.459 |
-0.371 |
| 33 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.453 |
3.278 |
| 34 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.446 |
0.121 |
| 35 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.444 |
3.552 |
| 36 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.444 |
3.009 |
| 37 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.442 |
1.376 |
| 38 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.439 |
5.795 |
| 39 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
0.436 |
-0.308 |
| 40 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
0.436 |
1.649 |
| 41 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.434 |
-1.859 |
| 42 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.433 |
-1.097 |
| 43 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.433 |
5.647 |
| 44 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.432 |
-1.743 |
| 45 |
267459_at |
AT2G33850
|
unknown |
0.430 |
-1.719 |
| 46 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.429 |
2.897 |
| 47 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.424 |
1.599 |
| 48 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
0.420 |
-0.793 |
| 49 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.418 |
1.600 |
| 50 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.417 |
-2.551 |
| 51 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
0.415 |
-0.641 |
| 52 |
258480_at |
AT3G02640
|
unknown |
0.414 |
5.706 |
| 53 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.407 |
-1.304 |
| 54 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.406 |
0.130 |
| 55 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.402 |
-1.649 |
| 56 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.401 |
-0.757 |
| 57 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.400 |
1.662 |
| 58 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.399 |
3.489 |
| 59 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.399 |
-0.842 |
| 60 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.396 |
0.051 |
| 61 |
251142_at |
AT5G01015
|
unknown |
0.390 |
2.365 |
| 62 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
0.390 |
-1.008 |
| 63 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.387 |
0.121 |
| 64 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.385 |
3.455 |
| 65 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.384 |
1.365 |
| 66 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.382 |
1.281 |
| 67 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.381 |
-0.630 |
| 68 |
257512_at |
AT1G35250
|
ALT2, acyl-lipid thioesterase 2 |
0.381 |
0.121 |
| 69 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.381 |
1.843 |
| 70 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
0.379 |
-2.878 |
| 71 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.378 |
-0.411 |
| 72 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.378 |
1.794 |
| 73 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.375 |
-0.301 |
| 74 |
246415_at |
AT5G17160
|
unknown |
0.372 |
4.282 |
| 75 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.369 |
0.679 |
| 76 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.368 |
0.842 |
| 77 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
0.365 |
1.362 |
| 78 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
0.364 |
-0.359 |
| 79 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.363 |
2.570 |
| 80 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.361 |
5.983 |
| 81 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
0.360 |
2.439 |
| 82 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
0.358 |
6.044 |
| 83 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.357 |
1.237 |
| 84 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.356 |
-3.849 |
| 85 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.356 |
7.563 |
| 86 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.354 |
2.570 |
| 87 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
0.354 |
3.598 |
| 88 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.353 |
3.029 |
| 89 |
256125_at |
AT1G18250
|
ATLP-1 |
0.352 |
1.480 |
| 90 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.351 |
1.042 |
| 91 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
0.350 |
-2.187 |
| 92 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.350 |
0.121 |
| 93 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.349 |
5.453 |
| 94 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
0.344 |
1.511 |
| 95 |
249136_at |
AT5G43180
|
unknown |
0.343 |
2.520 |
| 96 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.343 |
2.008 |
| 97 |
267344_at |
AT2G44230
|
unknown |
0.341 |
5.916 |
| 98 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.341 |
0.121 |
| 99 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.339 |
3.391 |
| 100 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
0.338 |
1.292 |
| 101 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.337 |
1.246 |
| 102 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
0.333 |
-0.407 |
| 103 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.333 |
-0.814 |
| 104 |
266900_at |
AT2G34610
|
unknown |
0.332 |
-0.814 |
| 105 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
0.332 |
3.150 |
| 106 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.331 |
-0.141 |
| 107 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
0.331 |
1.045 |
| 108 |
248623_at |
AT5G49170
|
unknown |
0.331 |
3.269 |
| 109 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
0.330 |
1.975 |
| 110 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
0.330 |
4.371 |
| 111 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
0.327 |
-0.407 |
| 112 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.325 |
-0.407 |
| 113 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
0.325 |
0.473 |
| 114 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.325 |
0.654 |
| 115 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
0.322 |
3.132 |
| 116 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
0.321 |
2.391 |
| 117 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
0.320 |
0.746 |
| 118 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.318 |
0.121 |
| 119 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.317 |
2.263 |
| 120 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
0.317 |
0.121 |
| 121 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.316 |
2.698 |
| 122 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.314 |
1.590 |
| 123 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.310 |
0.786 |
| 124 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.310 |
-0.019 |
| 125 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.308 |
3.896 |
| 126 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
0.306 |
-1.400 |
| 127 |
261657_at |
AT1G50050
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.305 |
0.121 |
| 128 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.305 |
5.078 |
| 129 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.304 |
-1.173 |
| 130 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
0.303 |
-0.156 |
| 131 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.303 |
-1.695 |
| 132 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.303 |
4.441 |
| 133 |
252661_at |
AT3G44450
|
unknown |
0.303 |
3.070 |
| 134 |
261691_at |
AT1G50060
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.303 |
0.121 |
| 135 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.302 |
0.458 |
| 136 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.301 |
0.876 |
| 137 |
261221_at |
AT1G19960
|
unknown |
0.300 |
1.601 |
| 138 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.300 |
-0.095 |
| 139 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.298 |
0.514 |
| 140 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.298 |
-0.957 |
| 141 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.297 |
-1.121 |
| 142 |
259891_at |
AT1G72730
|
[DEA(D/H)-box RNA helicase family protein] |
0.297 |
1.028 |
| 143 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.296 |
-1.097 |
| 144 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.296 |
0.449 |
| 145 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.295 |
2.075 |
| 146 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
0.292 |
-0.083 |
| 147 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.292 |
-0.365 |
| 148 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.292 |
2.733 |
| 149 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
0.291 |
2.726 |
| 150 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.290 |
2.360 |
| 151 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.290 |
3.889 |
| 152 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.289 |
2.148 |
| 153 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
0.289 |
0.769 |
| 154 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.289 |
-0.793 |
| 155 |
253740_at |
AT4G28706
|
[pfkB-like carbohydrate kinase family protein] |
0.288 |
0.253 |
| 156 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.286 |
0.833 |
| 157 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
0.286 |
1.474 |
| 158 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
0.286 |
0.121 |
| 159 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.286 |
0.649 |
| 160 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
0.286 |
-2.060 |
| 161 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.285 |
-5.270 |
| 162 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.284 |
0.121 |
| 163 |
260582_at |
AT2G47200
|
unknown |
0.283 |
0.121 |
| 164 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
0.281 |
0.018 |
| 165 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.281 |
-0.048 |
| 166 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
0.281 |
-1.442 |
| 167 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.281 |
2.584 |
| 168 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.280 |
3.101 |
| 169 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
0.280 |
-0.552 |
| 170 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
0.279 |
0.416 |
| 171 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
0.279 |
1.399 |
| 172 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.278 |
3.275 |
| 173 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.278 |
-1.184 |
| 174 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.277 |
-1.488 |
| 175 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.277 |
3.107 |
| 176 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.277 |
-2.155 |
| 177 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
0.276 |
5.999 |
| 178 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
0.276 |
-0.106 |
| 179 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.275 |
-0.010 |
| 180 |
264801_at |
AT1G08840
|
emb2411, embryo defective 2411 |
0.275 |
1.138 |
| 181 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
0.274 |
0.624 |
| 182 |
251778_at |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
0.273 |
7.366 |
| 183 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
0.273 |
7.267 |
| 184 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.273 |
-3.889 |
| 185 |
245353_at |
AT4G16000
|
unknown |
0.273 |
2.034 |
| 186 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.272 |
1.704 |
| 187 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
0.272 |
-0.793 |
| 188 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
0.271 |
0.027 |
| 189 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.271 |
0.654 |
| 190 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.271 |
1.262 |
| 191 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.270 |
6.159 |
| 192 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.269 |
-3.104 |
| 193 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
0.268 |
2.550 |
| 194 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
0.268 |
2.694 |
| 195 |
256122_at |
AT1G18180
|
[FUNCTIONS IN: oxidoreductase activity, acting on the CH-CH group of donors] |
0.268 |
-0.698 |
| 196 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.266 |
0.121 |
| 197 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.266 |
0.121 |
| 198 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
0.265 |
-4.542 |
| 199 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.265 |
2.919 |
| 200 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.265 |
4.365 |
| 201 |
254522_at |
AT4G19980
|
unknown |
0.264 |
0.121 |
| 202 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.264 |
0.096 |
| 203 |
254954_at |
AT4G10910
|
unknown |
0.264 |
1.600 |
| 204 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.263 |
0.620 |
| 205 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
0.262 |
-0.103 |
| 206 |
252089_at |
AT3G52110
|
unknown |
0.262 |
1.587 |
| 207 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.261 |
0.121 |
| 208 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
0.258 |
0.038 |
| 209 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.258 |
2.521 |
| 210 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.256 |
0.969 |
| 211 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
0.255 |
0.449 |
| 212 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
0.255 |
1.918 |
| 213 |
259979_at |
AT1G76600
|
unknown |
0.255 |
3.509 |
| 214 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.254 |
0.303 |
| 215 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.254 |
3.201 |
| 216 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
0.253 |
-0.582 |
| 217 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.253 |
3.647 |
| 218 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
0.253 |
-0.717 |
| 219 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
0.252 |
-0.147 |
| 220 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
0.252 |
-0.402 |
| 221 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.252 |
2.163 |
| 222 |
252412_at |
AT3G47295
|
unknown |
0.251 |
0.002 |
| 223 |
258177_at |
AT3G21660
|
[UBX domain-containing protein] |
0.250 |
-1.809 |
| 224 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
0.250 |
0.453 |
| 225 |
250223_at |
AT5G14070
|
ROXY2 |
0.249 |
-1.631 |
| 226 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.249 |
1.559 |
| 227 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.249 |
1.390 |
| 228 |
262922_at |
AT1G79420
|
unknown |
0.249 |
0.777 |
| 229 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.249 |
3.265 |
| 230 |
246584_at |
AT5G14730
|
unknown |
0.249 |
2.832 |
| 231 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.247 |
3.392 |
| 232 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.247 |
2.432 |
| 233 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.245 |
4.347 |
| 234 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.244 |
3.787 |
| 235 |
251503_at |
AT3G59140
|
ABCC10, ATP-binding cassette C10, ATMRP14, multidrug resistance-associated protein 14, MRP14, multidrug resistance-associated protein 14 |
0.244 |
2.742 |
| 236 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.244 |
0.401 |
| 237 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
0.244 |
-1.354 |
| 238 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.243 |
-1.108 |
| 239 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
0.243 |
0.121 |
| 240 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.243 |
-1.780 |
| 241 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.242 |
0.121 |
| 242 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
0.242 |
2.212 |
| 243 |
251920_at |
AT3G53900
|
PYRR, PYRIMIDINE R, UPP, uracil phosphoribosyltransferase |
0.242 |
-0.123 |
| 244 |
250837_at |
AT5G04620
|
ATBIOF, biotin F, BIO4, biotin 4, BIOF, biotin F |
0.242 |
1.349 |
| 245 |
245040_at |
AT2G26520
|
unknown |
0.241 |
3.194 |
| 246 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
0.241 |
1.076 |
| 247 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
0.240 |
3.124 |
| 248 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.240 |
6.978 |
| 249 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.239 |
3.639 |
| 250 |
250438_at |
AT5G10580
|
unknown |
0.237 |
0.654 |
| 251 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
0.237 |
6.715 |
| 252 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.236 |
0.055 |
| 253 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
0.236 |
-1.831 |
| 254 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
0.236 |
3.450 |
| 255 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
0.236 |
-0.337 |
| 256 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
0.236 |
0.121 |
| 257 |
266598_at |
AT2G46110
|
KPHMT1, ketopantoate hydroxymethyltransferase 1, PANB1 |
0.236 |
2.315 |
| 258 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
0.235 |
0.281 |
| 259 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.235 |
1.249 |
| 260 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.235 |
-0.260 |
| 261 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
0.235 |
-0.206 |
| 262 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.235 |
1.277 |
| 263 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.234 |
0.269 |
| 264 |
261344_at |
AT1G79710
|
[Major facilitator superfamily protein] |
0.234 |
4.067 |
| 265 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.234 |
1.373 |
| 266 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.233 |
6.576 |
| 267 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.233 |
1.217 |
| 268 |
245569_at |
AT4G14660
|
NRPE7 |
0.233 |
0.779 |
| 269 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
0.233 |
1.807 |
| 270 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-1.117 |
-11.007 |
| 271 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-1.042 |
-10.583 |
| 272 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-1.023 |
-10.462 |
| 273 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.945 |
-10.604 |
| 274 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.923 |
-4.688 |
| 275 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.867 |
-8.946 |
| 276 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.822 |
-10.909 |
| 277 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.759 |
-9.107 |
| 278 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
-0.706 |
-5.300 |
| 279 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.691 |
0.121 |
| 280 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.680 |
-1.874 |
| 281 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.678 |
-9.149 |
| 282 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.662 |
-6.792 |
| 283 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.649 |
-6.852 |
| 284 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.643 |
-6.156 |
| 285 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.637 |
-5.949 |
| 286 |
254263_at |
AT4G23493
|
unknown |
-0.634 |
-5.095 |
| 287 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
-0.633 |
-8.111 |
| 288 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.623 |
-10.084 |
| 289 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.614 |
-7.680 |
| 290 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.565 |
-10.703 |
| 291 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.565 |
1.439 |
| 292 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.556 |
-7.309 |
| 293 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.556 |
-7.737 |
| 294 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.556 |
-4.789 |
| 295 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.522 |
-8.332 |
| 296 |
244901_at |
ATMG00640
|
ORF25 |
-0.515 |
-6.148 |
| 297 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.510 |
-7.101 |
| 298 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.508 |
-8.872 |
| 299 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.502 |
-2.187 |
| 300 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
-0.502 |
-2.047 |
| 301 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
-0.490 |
-3.276 |
| 302 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.486 |
-8.393 |
| 303 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.481 |
-0.407 |
| 304 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.468 |
-1.560 |
| 305 |
260004_at |
AT1G67860
|
unknown |
-0.464 |
-1.091 |
| 306 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.449 |
0.866 |
| 307 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
-0.448 |
-1.218 |
| 308 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
-0.442 |
-6.873 |
| 309 |
264826_at |
AT1G03410
|
2A6 |
-0.441 |
-0.111 |
| 310 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.441 |
-6.787 |
| 311 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.440 |
0.114 |
| 312 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.439 |
0.121 |
| 313 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
-0.438 |
-4.447 |
| 314 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.433 |
1.640 |
| 315 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.426 |
0.633 |
| 316 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.426 |
2.127 |
| 317 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.419 |
-5.759 |
| 318 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.419 |
-0.022 |
| 319 |
264483_at |
AT1G77230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.418 |
-1.798 |
| 320 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.415 |
2.016 |
| 321 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.412 |
-1.162 |
| 322 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.412 |
-6.790 |
| 323 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.412 |
-2.873 |
| 324 |
259489_at |
AT1G15790
|
unknown |
-0.412 |
-0.047 |
| 325 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
-0.409 |
-5.770 |
| 326 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.409 |
0.121 |
| 327 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
-0.408 |
-5.050 |
| 328 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
-0.408 |
-4.728 |
| 329 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.404 |
0.965 |
| 330 |
256518_at |
AT1G66080
|
unknown |
-0.403 |
-4.938 |
| 331 |
260012_at |
AT1G67865
|
unknown |
-0.396 |
-0.266 |
| 332 |
256603_at |
AT3G28270
|
unknown |
-0.394 |
-2.691 |
| 333 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.392 |
0.348 |
| 334 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
-0.390 |
2.140 |
| 335 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
-0.389 |
-6.755 |
| 336 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
-0.384 |
0.120 |
| 337 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.383 |
-5.401 |
| 338 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.373 |
-0.207 |
| 339 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.372 |
3.980 |
| 340 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.371 |
-0.518 |
| 341 |
264636_at |
AT1G65490
|
unknown |
-0.371 |
-2.471 |
| 342 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.367 |
0.267 |
| 343 |
252888_at |
AT4G39210
|
APL3 |
-0.365 |
1.176 |
| 344 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
-0.361 |
-7.197 |
| 345 |
266707_at |
AT2G03310
|
unknown |
-0.361 |
-1.458 |
| 346 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.360 |
-1.153 |
| 347 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.356 |
2.192 |
| 348 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.348 |
-0.068 |
| 349 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
-0.348 |
1.746 |
| 350 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.343 |
0.762 |
| 351 |
247800_at |
AT5G58570
|
unknown |
-0.343 |
1.760 |
| 352 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.342 |
0.514 |
| 353 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
-0.340 |
-7.601 |
| 354 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.336 |
-1.676 |
| 355 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.335 |
0.121 |
| 356 |
257322_at |
ATMG01180
|
ORF111B |
-0.331 |
-3.970 |
| 357 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
-0.331 |
-5.641 |
| 358 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.328 |
0.121 |
| 359 |
258990_at |
AT3G08840
|
[D-alanine--D-alanine ligase family] |
-0.327 |
0.169 |
| 360 |
262014_at |
AT1G35660
|
unknown |
-0.326 |
-3.615 |
| 361 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.324 |
1.962 |
| 362 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
-0.324 |
-5.996 |
| 363 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.321 |
-6.427 |
| 364 |
264255_at |
AT1G09140
|
At-SR30, Serine/Arginine-Rich Protein Splicing Factor 30, ATSRP30, SERINE-ARGININE PROTEIN 30, ATSRP30.1, ATSRP30.2, SR30, Serine/Arginine-Rich Protein Splicing Factor 30 |
-0.320 |
-3.894 |
| 365 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.318 |
-2.305 |
| 366 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
-0.316 |
0.589 |
| 367 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.315 |
-1.231 |
| 368 |
248774_at |
AT5G47830
|
unknown |
-0.314 |
-5.972 |
| 369 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.313 |
-11.124 |
| 370 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.312 |
-5.430 |
| 371 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.311 |
-0.214 |
| 372 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.310 |
-0.092 |
| 373 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.309 |
-0.510 |
| 374 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.306 |
0.898 |
| 375 |
261505_at |
AT1G71696
|
SOL1, SUPPRESSOR OF LLP1 1 |
-0.303 |
0.586 |
| 376 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.301 |
-1.369 |
| 377 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.301 |
-4.733 |
| 378 |
253689_at |
AT4G29770
|
unknown |
-0.299 |
-6.039 |
| 379 |
266548_at |
AT2G35210
|
AGD10, MEE28, MATERNAL EFFECT EMBRYO ARREST 28, RPA, root and pollen arfgap |
-0.297 |
0.172 |
| 380 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.295 |
-5.328 |
| 381 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
-0.294 |
-1.744 |
| 382 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.291 |
-1.688 |
| 383 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
-0.290 |
-6.109 |
| 384 |
258827_at |
AT3G07150
|
unknown |
-0.289 |
-8.836 |
| 385 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.289 |
-1.590 |
| 386 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.287 |
0.669 |
| 387 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.287 |
-2.474 |
| 388 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.286 |
0.121 |
| 389 |
244906_at |
ATMG00690
|
ORF240A |
-0.285 |
-7.779 |
| 390 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.285 |
2.436 |
| 391 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
-0.284 |
-0.760 |
| 392 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.282 |
-2.602 |
| 393 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
-0.282 |
-0.706 |
| 394 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.281 |
0.004 |
| 395 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.276 |
1.147 |
| 396 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.276 |
0.007 |
| 397 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.276 |
-1.545 |
| 398 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.275 |
-0.089 |
| 399 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.274 |
4.989 |
| 400 |
251558_at |
AT3G57810
|
[Cysteine proteinases superfamily protein] |
-0.272 |
-2.055 |
| 401 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.270 |
-1.659 |
| 402 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
-0.270 |
-0.518 |
| 403 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.266 |
-2.177 |
| 404 |
246125_at |
AT5G19875
|
unknown |
-0.265 |
-6.972 |
| 405 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.265 |
-0.962 |
| 406 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
-0.262 |
0.487 |
| 407 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.262 |
0.121 |
| 408 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.261 |
-0.161 |
| 409 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.260 |
0.010 |
| 410 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
-0.259 |
-3.967 |
| 411 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-0.257 |
-0.066 |
| 412 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.256 |
1.134 |
| 413 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.256 |
0.405 |
| 414 |
254848_at |
AT4G11960
|
PGRL1B, PGR5-like B |
-0.256 |
-1.834 |
| 415 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
-0.254 |
-3.343 |
| 416 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.253 |
0.932 |
| 417 |
254402_at |
AT4G21310
|
unknown |
-0.250 |
-2.716 |
| 418 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.250 |
-4.697 |
| 419 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.250 |
0.102 |
| 420 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
-0.249 |
-1.921 |
| 421 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
-0.248 |
0.164 |
| 422 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.244 |
-1.742 |
| 423 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
-0.244 |
-5.595 |
| 424 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.243 |
-0.158 |
| 425 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.243 |
-2.603 |
| 426 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
-0.243 |
-0.681 |
| 427 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
-0.242 |
-1.814 |
| 428 |
265885_at |
AT2G42330
|
[GC-rich sequence DNA-binding factor-like protein with Tuftelin interacting domain] |
-0.242 |
-0.817 |
| 429 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.241 |
-2.834 |
| 430 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.240 |
0.844 |
| 431 |
250660_at |
AT5G07060
|
MAC5C, MOS4-associated complex subunit 5C |
-0.239 |
-1.391 |
| 432 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
-0.237 |
-0.543 |
| 433 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.237 |
0.968 |
| 434 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.237 |
-0.468 |
| 435 |
247731_at |
AT5G59590
|
UGT76E2, UDP-glucosyl transferase 76E2 |
-0.235 |
0.121 |
| 436 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
-0.235 |
-0.466 |
| 437 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
-0.233 |
-1.215 |
| 438 |
248172_at |
AT5G54660
|
[HSP20-like chaperones superfamily protein] |
-0.233 |
0.760 |
| 439 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.233 |
1.473 |
| 440 |
254693_at |
AT4G17880
|
MYC4 |
-0.232 |
-1.042 |
| 441 |
249147_at |
AT5G43330
|
c-NAD-MDH2, cytosolic-NAD-dependent malate dehydrogenase 2 |
-0.232 |
-3.318 |
| 442 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
-0.231 |
-4.518 |
| 443 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.231 |
-1.010 |
| 444 |
248609_at |
AT5G49440
|
unknown |
-0.229 |
-0.978 |
| 445 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
-0.229 |
-2.918 |
| 446 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
-0.228 |
-6.702 |
| 447 |
257334_at |
ATMG01370
|
ORF111D |
-0.226 |
-4.859 |
| 448 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
-0.226 |
2.645 |
| 449 |
259666_at |
AT1G55310
|
At-SCL33, SC35-LIKE SPLICING FACTOR 33, ATSCL33, SC35-like splicing factor 33, SCL33, SC35-LIKE SPLICING FACTOR 33, SR33, SC35-like splicing factor 33 |
-0.225 |
-2.591 |
| 450 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.225 |
-3.913 |
| 451 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.225 |
-1.000 |
| 452 |
265330_at |
AT2G18440
|
GUT15, GENE WITH UNSTABLE TRANSCRIPT 15 |
-0.224 |
-0.183 |
| 453 |
256574_at |
AT3G14780
|
unknown |
-0.223 |
-2.039 |
| 454 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
-0.222 |
-0.247 |
| 455 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
-0.222 |
-1.502 |
| 456 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.222 |
-2.200 |
| 457 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.222 |
-0.170 |
| 458 |
250013_at |
AT5G18040
|
unknown |
-0.221 |
-1.850 |
| 459 |
253854_at |
AT4G27900
|
[CCT motif family protein] |
-0.221 |
-0.628 |
| 460 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.220 |
-1.236 |
| 461 |
253351_at |
AT4G33700
|
unknown |
-0.220 |
0.452 |
| 462 |
260062_at |
AT1G73710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.220 |
-2.383 |
| 463 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.220 |
-10.154 |
| 464 |
254320_at |
AT4G22580
|
[Exostosin family protein] |
-0.219 |
0.603 |
| 465 |
247918_at |
AT5G57610
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.219 |
-2.164 |
| 466 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
-0.218 |
1.291 |
| 467 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.218 |
2.600 |
| 468 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.218 |
-1.045 |
| 469 |
249201_at |
AT5G42370
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.218 |
-1.557 |
| 470 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.218 |
0.932 |
| 471 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.217 |
0.415 |
| 472 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
-0.217 |
-4.627 |
| 473 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.216 |
0.813 |
| 474 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.216 |
-1.988 |
| 475 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
-0.216 |
-0.211 |
| 476 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
-0.215 |
-0.377 |
| 477 |
259091_at |
AT3G04890
|
[Uncharacterized conserved protein (DUF2358)] |
-0.215 |
0.095 |
| 478 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
-0.215 |
-3.690 |
| 479 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
-0.214 |
-0.815 |
| 480 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.214 |
-0.642 |
| 481 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.214 |
1.678 |
| 482 |
252885_at |
AT4G39260
|
ATGRP8, GLYCINE-RICH PROTEIN 8, CCR1, cold, circadian rhythm, and RNA binding 1, GR-RBP8, glycine-rich RNA-binding protein 8, GRP8 |
-0.213 |
1.687 |
| 483 |
257517_at |
AT3G16330
|
unknown |
-0.213 |
0.566 |
| 484 |
252233_at |
AT3G49690
|
ATMYB84, MYB DOMAIN PROTEIN 84, MYB84, myb domain protein 84, RAX3, REGULATOR OF AXILLARY MERISTEMS3 |
-0.212 |
-1.348 |
| 485 |
254457_at |
AT4G21170
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.212 |
-0.951 |
| 486 |
248248_at |
AT5G53120
|
ATSPDS3, SPERMIDINE SYNTHASE 3, SPDS3, spermidine synthase 3, SPMS |
-0.212 |
-2.005 |
| 487 |
263921_at |
AT2G36460
|
FBA6, fructose-bisphosphate aldolase 6 |
-0.211 |
-4.973 |
| 488 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.211 |
-1.399 |
| 489 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.211 |
0.669 |
| 490 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
-0.210 |
-0.568 |
| 491 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
-0.210 |
-0.700 |
| 492 |
258019_at |
AT3G19470
|
[F-box and associated interaction domains-containing protein] |
-0.209 |
0.883 |
| 493 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.209 |
-1.148 |
| 494 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.208 |
0.917 |
| 495 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.208 |
-1.541 |
| 496 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.207 |
1.529 |
| 497 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.206 |
-2.048 |
| 498 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.206 |
0.121 |
| 499 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
-0.206 |
-3.386 |
| 500 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.205 |
0.359 |
| 501 |
247044_at |
AT5G66850
|
MAPKKK5, mitogen-activated protein kinase kinase kinase 5 |
-0.205 |
-1.514 |
| 502 |
263146_at |
AT1G53940
|
GLIP2, GDSL-motif lipase 2 |
-0.204 |
0.121 |
| 503 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.204 |
-1.213 |
| 504 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.204 |
-0.300 |
| 505 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
-0.203 |
-1.977 |
| 506 |
246888_at |
AT5G26270
|
unknown |
-0.203 |
-0.899 |
| 507 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.203 |
-0.068 |
| 508 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.202 |
-0.756 |
| 509 |
253199_at |
AT4G35410
|
[Clathrin adaptor complex small chain family protein] |
-0.201 |
1.294 |
| 510 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.201 |
1.180 |
| 511 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.201 |
0.431 |
| 512 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.201 |
-2.152 |
| 513 |
265719_at |
AT2G03500
|
[Homeodomain-like superfamily protein] |
-0.200 |
1.608 |
| 514 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.200 |
1.041 |
| 515 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
-0.200 |
-0.719 |
| 516 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.200 |
0.270 |
| 517 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
-0.199 |
-1.170 |
| 518 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
-0.199 |
2.459 |
| 519 |
255760_at |
AT1G16780
|
AtVHP2;2, VHP2;2 |
-0.199 |
-0.080 |
| 520 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
-0.199 |
-1.313 |
| 521 |
248445_at |
AT5G51170
|
unknown |
-0.198 |
-1.830 |
| 522 |
258460_at |
AT3G17330
|
ECT6, evolutionarily conserved C-terminal region 6 |
-0.198 |
-2.075 |
| 523 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.198 |
0.595 |
| 524 |
259162_at |
AT3G01640
|
ATGLCAK, ARABIDOPSIS THALIANA GLUCURONOKINASE, GLCAK, glucuronokinase G |
-0.198 |
0.347 |
| 525 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.197 |
3.541 |
| 526 |
249122_at |
AT5G43850
|
ARD4, ATARD4 |
-0.197 |
-2.449 |
| 527 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.196 |
-0.389 |
| 528 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.196 |
-1.183 |
| 529 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.196 |
-1.459 |
| 530 |
252469_at |
AT3G46920
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.195 |
-0.875 |
| 531 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.195 |
0.381 |
| 532 |
262291_at |
AT1G70790
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.195 |
-0.732 |
| 533 |
247504_at |
AT5G61990
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.195 |
-0.947 |
| 534 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.195 |
0.402 |
| 535 |
249000_at |
AT5G44980
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.194 |
0.466 |
| 536 |
253020_at |
AT4G38020
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
-0.194 |
-2.350 |
| 537 |
245618_at |
AT4G14510
|
ATCFM3B, CFM3B, CRM family member 3B |
-0.194 |
-0.677 |