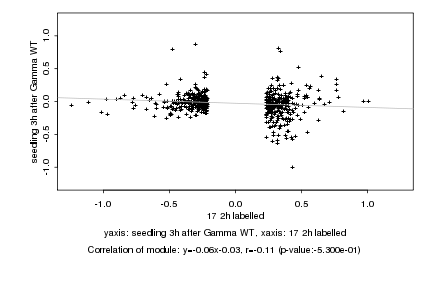
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
17 2h labelled |
Log2 signal ratio
seedling 3h after Gamma WT |
| 1 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.008 |
0.005 |
| 2 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.967 |
0.003 |
| 3 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.817 |
-0.140 |
| 4 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.777 |
0.062 |
| 5 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.761 |
0.336 |
| 6 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.761 |
0.182 |
| 7 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.760 |
0.273 |
| 8 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.710 |
-0.009 |
| 9 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
0.670 |
-0.032 |
| 10 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.646 |
0.394 |
| 11 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.637 |
0.055 |
| 12 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.631 |
0.045 |
| 13 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.625 |
-0.279 |
| 14 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.623 |
0.173 |
| 15 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.599 |
-0.029 |
| 16 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.566 |
0.239 |
| 17 |
261247_at |
AT1G20070
|
unknown |
0.554 |
0.200 |
| 18 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.552 |
-0.080 |
| 19 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.546 |
0.073 |
| 20 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
0.543 |
-0.458 |
| 21 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.540 |
0.070 |
| 22 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.533 |
0.254 |
| 23 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
0.532 |
0.099 |
| 24 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.521 |
-0.152 |
| 25 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.520 |
0.078 |
| 26 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.517 |
0.050 |
| 27 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
0.501 |
-0.056 |
| 28 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.487 |
-0.260 |
| 29 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
0.474 |
-0.050 |
| 30 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.473 |
0.169 |
| 31 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.473 |
0.524 |
| 32 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.469 |
-0.202 |
| 33 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
0.465 |
0.117 |
| 34 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.465 |
0.081 |
| 35 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
0.457 |
-0.042 |
| 36 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.454 |
-0.531 |
| 37 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.454 |
-0.099 |
| 38 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.446 |
-0.045 |
| 39 |
267344_at |
AT2G44230
|
unknown |
0.440 |
-0.167 |
| 40 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.432 |
0.058 |
| 41 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.430 |
-0.989 |
| 42 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
0.427 |
-0.270 |
| 43 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
0.427 |
-0.573 |
| 44 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.426 |
-0.030 |
| 45 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.425 |
-0.129 |
| 46 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.423 |
0.279 |
| 47 |
258480_at |
AT3G02640
|
unknown |
0.420 |
-0.540 |
| 48 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
0.415 |
-0.188 |
| 49 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.414 |
0.065 |
| 50 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.411 |
0.138 |
| 51 |
262971_at |
AT1G75640
|
[Leucine-rich receptor-like protein kinase family protein] |
0.411 |
-0.259 |
| 52 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
0.407 |
-0.303 |
| 53 |
246585_at |
AT5G14750
|
ATMYB66, myb domain protein 66, MYB66, myb domain protein 66, WER, WEREWOLF, WER1, WEREWOLF 1 |
0.406 |
0.123 |
| 54 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.403 |
-0.117 |
| 55 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.401 |
-0.027 |
| 56 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.401 |
-0.162 |
| 57 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
0.399 |
-0.166 |
| 58 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
0.398 |
0.066 |
| 59 |
250223_at |
AT5G14070
|
ROXY2 |
0.398 |
-0.449 |
| 60 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.396 |
-0.250 |
| 61 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.395 |
0.090 |
| 62 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
0.395 |
-0.075 |
| 63 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.395 |
0.027 |
| 64 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
0.394 |
-0.345 |
| 65 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.394 |
-0.294 |
| 66 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.394 |
-0.153 |
| 67 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.393 |
0.011 |
| 68 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
0.392 |
0.025 |
| 69 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
0.390 |
-0.160 |
| 70 |
245353_at |
AT4G16000
|
unknown |
0.387 |
-0.445 |
| 71 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.385 |
0.076 |
| 72 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.384 |
-0.251 |
| 73 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.384 |
-0.059 |
| 74 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.384 |
-0.118 |
| 75 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.382 |
0.058 |
| 76 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.381 |
-0.559 |
| 77 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
0.381 |
0.032 |
| 78 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
0.380 |
-0.013 |
| 79 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.380 |
0.057 |
| 80 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.379 |
-0.028 |
| 81 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.379 |
-0.060 |
| 82 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
0.379 |
0.020 |
| 83 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.377 |
-0.091 |
| 84 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.377 |
0.018 |
| 85 |
262701_at |
AT1G16480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.375 |
0.099 |
| 86 |
267459_at |
AT2G33850
|
unknown |
0.374 |
0.133 |
| 87 |
251503_at |
AT3G59140
|
ABCC10, ATP-binding cassette C10, ATMRP14, multidrug resistance-associated protein 14, MRP14, multidrug resistance-associated protein 14 |
0.373 |
-0.019 |
| 88 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.373 |
-0.267 |
| 89 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.372 |
-0.506 |
| 90 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
0.371 |
-0.110 |
| 91 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.369 |
0.029 |
| 92 |
256125_at |
AT1G18250
|
ATLP-1 |
0.366 |
-0.356 |
| 93 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.365 |
0.006 |
| 94 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.364 |
0.249 |
| 95 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.361 |
0.248 |
| 96 |
264801_at |
AT1G08840
|
emb2411, embryo defective 2411 |
0.360 |
0.052 |
| 97 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.360 |
-0.026 |
| 98 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.359 |
-0.266 |
| 99 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.356 |
-0.252 |
| 100 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.354 |
0.004 |
| 101 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.354 |
-0.156 |
| 102 |
252412_at |
AT3G47295
|
unknown |
0.350 |
-0.060 |
| 103 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.349 |
-0.100 |
| 104 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.349 |
0.102 |
| 105 |
248623_at |
AT5G49170
|
unknown |
0.349 |
-0.002 |
| 106 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
0.348 |
0.047 |
| 107 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.346 |
-0.043 |
| 108 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.345 |
-0.101 |
| 109 |
262922_at |
AT1G79420
|
unknown |
0.345 |
-0.202 |
| 110 |
249887_at |
AT5G22310
|
unknown |
0.342 |
-0.014 |
| 111 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.342 |
-0.076 |
| 112 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.341 |
0.767 |
| 113 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.338 |
-0.175 |
| 114 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
0.337 |
-0.371 |
| 115 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.337 |
-0.065 |
| 116 |
254954_at |
AT4G10910
|
unknown |
0.336 |
-0.353 |
| 117 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.336 |
-0.071 |
| 118 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.335 |
0.188 |
| 119 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.333 |
-0.033 |
| 120 |
258867_at |
AT3G03130
|
unknown |
0.333 |
-0.397 |
| 121 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.333 |
-0.139 |
| 122 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.332 |
0.075 |
| 123 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
0.332 |
-0.010 |
| 124 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
0.331 |
-0.051 |
| 125 |
246584_at |
AT5G14730
|
unknown |
0.330 |
0.250 |
| 126 |
257551_at |
AT3G13270
|
unknown |
0.326 |
0.005 |
| 127 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
0.326 |
-0.220 |
| 128 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.326 |
-0.100 |
| 129 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
0.324 |
0.039 |
| 130 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.324 |
0.807 |
| 131 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
0.323 |
0.037 |
| 132 |
259968_at |
AT1G76530
|
[Auxin efflux carrier family protein] |
0.323 |
0.033 |
| 133 |
260801_at |
AT1G78430
|
RIP4, ROP interactive partner 4 |
0.322 |
-0.134 |
| 134 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.322 |
-0.258 |
| 135 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.322 |
0.108 |
| 136 |
266825_at |
AT2G22890
|
[Kua-ubiquitin conjugating enzyme hybrid localisation domain] |
0.320 |
-0.081 |
| 137 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.320 |
0.365 |
| 138 |
246505_at |
AT5G16250
|
unknown |
0.319 |
-0.503 |
| 139 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.319 |
0.023 |
| 140 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
0.317 |
-0.631 |
| 141 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.317 |
0.202 |
| 142 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
0.317 |
-0.043 |
| 143 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.316 |
-0.001 |
| 144 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.316 |
0.332 |
| 145 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.315 |
0.020 |
| 146 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.315 |
-0.288 |
| 147 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
0.314 |
0.377 |
| 148 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.314 |
-0.220 |
| 149 |
251249_at |
AT3G62160
|
[HXXXD-type acyl-transferase family protein] |
0.314 |
-0.045 |
| 150 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.314 |
-0.067 |
| 151 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.313 |
-0.148 |
| 152 |
248057_at |
AT5G55520
|
unknown |
0.313 |
-0.593 |
| 153 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.312 |
-0.048 |
| 154 |
248423_at |
AT5G51670
|
unknown |
0.308 |
-0.010 |
| 155 |
250909_at |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.307 |
-0.027 |
| 156 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
0.305 |
-0.318 |
| 157 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
0.305 |
-0.044 |
| 158 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.303 |
0.168 |
| 159 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.302 |
-0.165 |
| 160 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.302 |
0.123 |
| 161 |
255733_at |
AT1G25400
|
unknown |
0.299 |
0.016 |
| 162 |
261344_at |
AT1G79710
|
[Major facilitator superfamily protein] |
0.299 |
0.017 |
| 163 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
0.298 |
-0.360 |
| 164 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.298 |
-0.057 |
| 165 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.297 |
0.021 |
| 166 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
0.296 |
-0.163 |
| 167 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
0.296 |
0.060 |
| 168 |
259891_at |
AT1G72730
|
[DEA(D/H)-box RNA helicase family protein] |
0.296 |
-0.041 |
| 169 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.293 |
-0.196 |
| 170 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
0.293 |
-0.379 |
| 171 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.291 |
-0.087 |
| 172 |
259979_at |
AT1G76600
|
unknown |
0.291 |
0.060 |
| 173 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.288 |
-0.091 |
| 174 |
252089_at |
AT3G52110
|
unknown |
0.288 |
-0.463 |
| 175 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.288 |
-0.020 |
| 176 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.286 |
-0.053 |
| 177 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.286 |
-0.026 |
| 178 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
0.285 |
-0.312 |
| 179 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.284 |
0.117 |
| 180 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.282 |
0.212 |
| 181 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
0.282 |
-0.508 |
| 182 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.282 |
-0.086 |
| 183 |
254532_at |
AT4G19660
|
ATNPR4, NPR4, NPR1-like protein 4 |
0.281 |
-0.036 |
| 184 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.279 |
0.351 |
| 185 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
0.278 |
-0.116 |
| 186 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.278 |
-0.160 |
| 187 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.278 |
0.074 |
| 188 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.278 |
0.042 |
| 189 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.278 |
-0.043 |
| 190 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
0.277 |
0.102 |
| 191 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
0.277 |
-0.597 |
| 192 |
256799_at |
AT3G18560
|
unknown |
0.276 |
-0.374 |
| 193 |
256320_at |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
0.276 |
-0.052 |
| 194 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.276 |
-0.229 |
| 195 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.275 |
-0.121 |
| 196 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.274 |
-0.008 |
| 197 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
0.272 |
-0.046 |
| 198 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.271 |
-0.168 |
| 199 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
0.270 |
-0.223 |
| 200 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
0.269 |
-0.119 |
| 201 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.269 |
0.253 |
| 202 |
247585_at |
AT5G60680
|
unknown |
0.268 |
-0.148 |
| 203 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
0.268 |
-0.022 |
| 204 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.267 |
0.156 |
| 205 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.267 |
-0.088 |
| 206 |
247425_at |
AT5G62550
|
unknown |
0.267 |
-0.249 |
| 207 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.267 |
-0.082 |
| 208 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.265 |
0.027 |
| 209 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.264 |
0.028 |
| 210 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.263 |
-0.017 |
| 211 |
251142_at |
AT5G01015
|
unknown |
0.262 |
-0.057 |
| 212 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
0.262 |
-0.131 |
| 213 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.261 |
-0.113 |
| 214 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.260 |
-0.003 |
| 215 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
0.260 |
-0.395 |
| 216 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.260 |
-0.274 |
| 217 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.259 |
0.161 |
| 218 |
259298_at |
AT3G05370
|
AtRLP31, receptor like protein 31, RLP31, receptor like protein 31 |
0.259 |
0.206 |
| 219 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.258 |
0.064 |
| 220 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
0.258 |
-0.149 |
| 221 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
0.258 |
0.062 |
| 222 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
0.257 |
-0.094 |
| 223 |
259473_at |
AT1G19025
|
[DNA repair metallo-beta-lactamase family protein] |
0.257 |
-0.031 |
| 224 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.257 |
-0.058 |
| 225 |
254204_at |
AT4G24160
|
[alpha/beta-Hydrolases superfamily protein] |
0.257 |
0.112 |
| 226 |
260365_at |
AT1G70630
|
RAY1, REDUCED ARABINOSE YARIV 1 |
0.255 |
-0.078 |
| 227 |
258935_at |
AT3G10120
|
unknown |
0.255 |
0.072 |
| 228 |
252915_at |
AT4G38810
|
[Calcium-binding EF-hand family protein] |
0.254 |
-0.156 |
| 229 |
248981_at |
AT5G45110
|
ATNPR3, NPR3, NPR1-like protein 3 |
0.254 |
0.026 |
| 230 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.254 |
-0.094 |
| 231 |
246042_at |
AT5G19440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.254 |
-0.026 |
| 232 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.254 |
0.005 |
| 233 |
260602_at |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
0.254 |
0.119 |
| 234 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
0.254 |
0.054 |
| 235 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
0.254 |
-0.387 |
| 236 |
247287_at |
AT5G64230
|
unknown |
0.253 |
0.150 |
| 237 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
0.251 |
0.086 |
| 238 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.250 |
-0.022 |
| 239 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
0.249 |
0.012 |
| 240 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.249 |
0.017 |
| 241 |
260761_at |
AT1G49150
|
unknown |
0.246 |
0.013 |
| 242 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
0.245 |
0.020 |
| 243 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.245 |
-0.068 |
| 244 |
260362_at |
AT1G70530
|
CRK3, cysteine-rich RLK (RECEPTOR-like protein kinase) 3 |
0.245 |
0.207 |
| 245 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
0.244 |
-0.110 |
| 246 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.244 |
-0.085 |
| 247 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
0.243 |
-0.314 |
| 248 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
0.243 |
-0.023 |
| 249 |
249060_at |
AT5G44560
|
VPS2.2 |
0.243 |
-0.058 |
| 250 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.242 |
-0.010 |
| 251 |
256454_at |
AT1G75280
|
[NmrA-like negative transcriptional regulator family protein] |
0.242 |
0.083 |
| 252 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.241 |
-0.160 |
| 253 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.239 |
-0.085 |
| 254 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
0.238 |
0.142 |
| 255 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
0.238 |
-0.105 |
| 256 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
0.238 |
-0.079 |
| 257 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.238 |
-0.162 |
| 258 |
245166_at |
AT2G33170
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
0.237 |
-0.172 |
| 259 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.236 |
-0.097 |
| 260 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.235 |
-0.309 |
| 261 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
0.235 |
-0.196 |
| 262 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.235 |
-0.050 |
| 263 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
0.234 |
0.016 |
| 264 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.234 |
-0.243 |
| 265 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.234 |
0.036 |
| 266 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
0.234 |
-0.192 |
| 267 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.233 |
-0.096 |
| 268 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.232 |
-0.210 |
| 269 |
256409_at |
AT1G66620
|
[Protein with RING/U-box and TRAF-like domains] |
0.232 |
0.076 |
| 270 |
258105_at |
AT3G23605
|
[Ubiquitin-like superfamily protein] |
0.232 |
-0.000 |
| 271 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.231 |
0.137 |
| 272 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
0.231 |
-0.537 |
| 273 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-1.248 |
-0.051 |
| 274 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-1.118 |
-0.003 |
| 275 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-1.022 |
-0.154 |
| 276 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.979 |
0.041 |
| 277 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.973 |
-0.186 |
| 278 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.905 |
0.044 |
| 279 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.875 |
0.060 |
| 280 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
-0.844 |
0.106 |
| 281 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.782 |
-0.010 |
| 282 |
254263_at |
AT4G23493
|
unknown |
-0.775 |
-0.105 |
| 283 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.774 |
0.058 |
| 284 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.765 |
-0.060 |
| 285 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.709 |
0.099 |
| 286 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.678 |
-0.109 |
| 287 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.675 |
0.070 |
| 288 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.653 |
0.017 |
| 289 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.641 |
0.057 |
| 290 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.619 |
-0.216 |
| 291 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.610 |
-0.030 |
| 292 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.605 |
-0.103 |
| 293 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
-0.600 |
-0.039 |
| 294 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.578 |
0.119 |
| 295 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.547 |
-0.087 |
| 296 |
252888_at |
AT4G39210
|
APL3 |
-0.544 |
0.000 |
| 297 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.540 |
0.002 |
| 298 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
-0.529 |
0.262 |
| 299 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.528 |
-0.247 |
| 300 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.520 |
-0.091 |
| 301 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
-0.517 |
0.005 |
| 302 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.516 |
-0.078 |
| 303 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.497 |
-0.074 |
| 304 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.495 |
-0.182 |
| 305 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.494 |
-0.211 |
| 306 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.493 |
-0.183 |
| 307 |
244901_at |
ATMG00640
|
ORF25 |
-0.492 |
-0.074 |
| 308 |
264636_at |
AT1G65490
|
unknown |
-0.487 |
-0.183 |
| 309 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
-0.483 |
0.799 |
| 310 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.483 |
0.005 |
| 311 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.481 |
-0.116 |
| 312 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.480 |
0.009 |
| 313 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
-0.477 |
-0.103 |
| 314 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.473 |
-0.030 |
| 315 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.467 |
0.006 |
| 316 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
-0.459 |
-0.125 |
| 317 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.458 |
-0.023 |
| 318 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-0.452 |
0.006 |
| 319 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.448 |
-0.097 |
| 320 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.443 |
-0.038 |
| 321 |
247964_at |
AT5G56600
|
PFN3, PROFILIN 3, PRF3, profilin 3 |
-0.440 |
0.120 |
| 322 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.439 |
-0.028 |
| 323 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.439 |
0.122 |
| 324 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.439 |
0.084 |
| 325 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.437 |
-0.237 |
| 326 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.436 |
0.038 |
| 327 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.435 |
0.143 |
| 328 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.433 |
0.013 |
| 329 |
248774_at |
AT5G47830
|
unknown |
-0.430 |
-0.088 |
| 330 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.421 |
-0.127 |
| 331 |
256518_at |
AT1G66080
|
unknown |
-0.420 |
0.349 |
| 332 |
244931_at |
ATMG00630
|
ORF110B |
-0.418 |
0.040 |
| 333 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.414 |
-0.066 |
| 334 |
266707_at |
AT2G03310
|
unknown |
-0.413 |
-0.006 |
| 335 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.413 |
-0.007 |
| 336 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.409 |
-0.107 |
| 337 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
-0.403 |
-0.027 |
| 338 |
260872_at |
AT1G21350
|
[Thioredoxin superfamily protein] |
-0.395 |
0.041 |
| 339 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.392 |
-0.071 |
| 340 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.391 |
0.017 |
| 341 |
258990_at |
AT3G08840
|
[D-alanine--D-alanine ligase family] |
-0.388 |
0.105 |
| 342 |
254190_at |
AT4G23885
|
unknown |
-0.386 |
0.003 |
| 343 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.385 |
-0.003 |
| 344 |
264255_at |
AT1G09140
|
At-SR30, Serine/Arginine-Rich Protein Splicing Factor 30, ATSRP30, SERINE-ARGININE PROTEIN 30, ATSRP30.1, ATSRP30.2, SR30, Serine/Arginine-Rich Protein Splicing Factor 30 |
-0.379 |
-0.015 |
| 345 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.378 |
-0.191 |
| 346 |
247800_at |
AT5G58570
|
unknown |
-0.378 |
-0.011 |
| 347 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
-0.374 |
0.136 |
| 348 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
-0.373 |
-0.083 |
| 349 |
259489_at |
AT1G15790
|
unknown |
-0.369 |
0.118 |
| 350 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
-0.364 |
-0.008 |
| 351 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.364 |
-0.010 |
| 352 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.359 |
0.060 |
| 353 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.359 |
-0.115 |
| 354 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.359 |
-0.030 |
| 355 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.358 |
0.003 |
| 356 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.358 |
0.018 |
| 357 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
-0.357 |
0.132 |
| 358 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.355 |
-0.121 |
| 359 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
-0.354 |
0.111 |
| 360 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.352 |
-0.061 |
| 361 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.349 |
0.097 |
| 362 |
249147_at |
AT5G43330
|
c-NAD-MDH2, cytosolic-NAD-dependent malate dehydrogenase 2 |
-0.346 |
-0.002 |
| 363 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.345 |
0.176 |
| 364 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.342 |
0.070 |
| 365 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.341 |
-0.230 |
| 366 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
-0.337 |
-0.032 |
| 367 |
264483_at |
AT1G77230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.337 |
0.013 |
| 368 |
262162_at |
AT1G78020
|
unknown |
-0.335 |
-0.099 |
| 369 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.335 |
-0.147 |
| 370 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.334 |
-0.075 |
| 371 |
260444_at |
AT1G68300
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.333 |
0.108 |
| 372 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.332 |
-0.109 |
| 373 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.330 |
-0.050 |
| 374 |
246125_at |
AT5G19875
|
unknown |
-0.330 |
0.137 |
| 375 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.328 |
0.054 |
| 376 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.325 |
-0.076 |
| 377 |
264826_at |
AT1G03410
|
2A6 |
-0.324 |
0.099 |
| 378 |
263921_at |
AT2G36460
|
FBA6, fructose-bisphosphate aldolase 6 |
-0.321 |
0.007 |
| 379 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
-0.319 |
0.045 |
| 380 |
248248_at |
AT5G53120
|
ATSPDS3, SPERMIDINE SYNTHASE 3, SPDS3, spermidine synthase 3, SPMS |
-0.318 |
-0.036 |
| 381 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.316 |
0.122 |
| 382 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.315 |
0.037 |
| 383 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.314 |
0.062 |
| 384 |
250016_at |
AT5G18100
|
CSD3, copper/zinc superoxide dismutase 3 |
-0.310 |
0.000 |
| 385 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
-0.309 |
-0.010 |
| 386 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.309 |
-0.088 |
| 387 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.308 |
0.877 |
| 388 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
-0.307 |
0.222 |
| 389 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
-0.307 |
0.262 |
| 390 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.305 |
-0.205 |
| 391 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.305 |
-0.081 |
| 392 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
-0.305 |
-0.046 |
| 393 |
257322_at |
ATMG01180
|
ORF111B |
-0.305 |
-0.004 |
| 394 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.303 |
-0.020 |
| 395 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
-0.303 |
0.073 |
| 396 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.301 |
-0.199 |
| 397 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.301 |
0.015 |
| 398 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
-0.296 |
0.084 |
| 399 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.296 |
0.206 |
| 400 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
-0.295 |
0.057 |
| 401 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.295 |
0.024 |
| 402 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.293 |
0.185 |
| 403 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.292 |
-0.004 |
| 404 |
249122_at |
AT5G43850
|
ARD4, ATARD4 |
-0.292 |
0.003 |
| 405 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.290 |
-0.067 |
| 406 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.290 |
-0.140 |
| 407 |
261641_at |
AT1G27670
|
unknown |
-0.286 |
-0.116 |
| 408 |
245341_at |
AT4G16447
|
unknown |
-0.286 |
0.126 |
| 409 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.283 |
-0.099 |
| 410 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
-0.283 |
0.107 |
| 411 |
260714_at |
AT1G14980
|
CPN10, chaperonin 10 |
-0.282 |
-0.005 |
| 412 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.282 |
0.047 |
| 413 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.281 |
-0.040 |
| 414 |
252946_at |
AT4G39235
|
unknown |
-0.280 |
0.176 |
| 415 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.280 |
-0.080 |
| 416 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.279 |
0.118 |
| 417 |
266848_at |
AT2G25950
|
unknown |
-0.278 |
0.127 |
| 418 |
251011_at |
AT5G02560
|
HTA12, histone H2A 12 |
-0.277 |
0.051 |
| 419 |
244906_at |
ATMG00690
|
ORF240A |
-0.275 |
-0.056 |
| 420 |
247659_at |
AT5G60040
|
NRPC1, nuclear RNA polymerase C1 |
-0.274 |
0.046 |
| 421 |
264814_at |
AT2G17900
|
ASHR1, ASH1-related 1, SDG37, SET domain group 37 |
-0.274 |
0.096 |
| 422 |
254624_at |
AT4G18580
|
unknown |
-0.273 |
0.091 |
| 423 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.273 |
-0.111 |
| 424 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.273 |
0.097 |
| 425 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.272 |
-0.019 |
| 426 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
-0.271 |
0.021 |
| 427 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-0.270 |
-0.067 |
| 428 |
265510_at |
AT2G05630
|
ATG8D |
-0.269 |
-0.010 |
| 429 |
254848_at |
AT4G11960
|
PGRL1B, PGR5-like B |
-0.268 |
0.035 |
| 430 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.268 |
-0.030 |
| 431 |
249978_at |
AT5G18850
|
unknown |
-0.264 |
-0.047 |
| 432 |
248258_at |
AT5G53400
|
BOB1, BOBBER1 |
-0.263 |
-0.029 |
| 433 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.263 |
0.096 |
| 434 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.263 |
0.032 |
| 435 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.263 |
0.006 |
| 436 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.262 |
0.100 |
| 437 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.262 |
-0.162 |
| 438 |
259299_at |
AT3G05080
|
unknown |
-0.262 |
0.031 |
| 439 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.261 |
-0.031 |
| 440 |
245243_at |
AT1G44414
|
unknown |
-0.261 |
0.046 |
| 441 |
244923_s_at |
ATMG01020
|
ORF153B |
-0.260 |
-0.088 |
| 442 |
251558_at |
AT3G57810
|
[Cysteine proteinases superfamily protein] |
-0.259 |
0.000 |
| 443 |
253854_at |
AT4G27900
|
[CCT motif family protein] |
-0.258 |
-0.046 |
| 444 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.258 |
0.029 |
| 445 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.258 |
-0.062 |
| 446 |
263296_at |
AT2G38800
|
[Plant calmodulin-binding protein-related] |
-0.258 |
-0.007 |
| 447 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
-0.257 |
0.021 |
| 448 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.257 |
-0.091 |
| 449 |
253351_at |
AT4G33700
|
unknown |
-0.256 |
-0.033 |
| 450 |
261013_at |
AT1G26440
|
ATUPS5, ureide permease 5, UPS5, UPS5, ureide permease 5 |
-0.255 |
0.028 |
| 451 |
264224_at |
AT1G67440
|
emb1688, embryo defective 1688 |
-0.255 |
0.060 |
| 452 |
247055_at |
AT5G66740
|
unknown |
-0.255 |
0.158 |
| 453 |
262313_at |
AT1G70900
|
unknown |
-0.255 |
-0.048 |
| 454 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.254 |
0.149 |
| 455 |
267484_at |
AT2G02760
|
ATUBC2, ubiquiting-conjugating enzyme 2, UBC2, ubiquitin-conjugating enzyme 2, UBC2, ubiquiting-conjugating enzyme 2 |
-0.253 |
0.049 |
| 456 |
261919_at |
AT1G65980
|
TPX1, thioredoxin-dependent peroxidase 1 |
-0.253 |
-0.016 |
| 457 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.253 |
-0.018 |
| 458 |
255412_at |
AT4G02980
|
ABP, ABP1, endoplasmic reticulum auxin binding protein 1 |
-0.253 |
0.058 |
| 459 |
267428_at |
AT2G34840
|
[Coatomer epsilon subunit] |
-0.252 |
0.139 |
| 460 |
259849_at |
AT1G72190
|
[D-isomer specific 2-hydroxyacid dehydrogenase family protein] |
-0.251 |
-0.077 |
| 461 |
253391_at |
AT4G32590
|
[2Fe-2S ferredoxin-like superfamily protein] |
-0.250 |
0.010 |
| 462 |
258782_at |
AT3G11750
|
FOLB1 |
-0.249 |
-0.160 |
| 463 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.249 |
0.077 |
| 464 |
263369_at |
AT2G20480
|
unknown |
-0.249 |
-0.063 |
| 465 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.247 |
-0.107 |
| 466 |
262388_at |
AT1G49320
|
unknown |
-0.247 |
-0.078 |
| 467 |
262578_at |
AT1G15300
|
CYTOSLEEPER, CYTOSLEEPER |
-0.247 |
-0.015 |
| 468 |
260350_at |
AT1G69410
|
ATELF5A-3, EUKARYOTIC ELONGATION FACTOR 5A-3, ELF5A-3, eukaryotic elongation factor 5A-3 |
-0.247 |
0.025 |
| 469 |
258576_at |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
-0.247 |
0.041 |
| 470 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
-0.246 |
-0.014 |
| 471 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.245 |
-0.124 |
| 472 |
249441_at |
AT5G39730
|
[AIG2-like (avirulence induced gene) family protein] |
-0.244 |
0.007 |
| 473 |
253020_at |
AT4G38020
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
-0.243 |
0.076 |
| 474 |
259089_at |
AT3G04960
|
unknown |
-0.243 |
0.033 |
| 475 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.243 |
-0.041 |
| 476 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
-0.242 |
0.105 |
| 477 |
259551_at |
AT1G21190
|
LSM3A, SM-like 3A |
-0.241 |
0.073 |
| 478 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
-0.241 |
0.183 |
| 479 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
-0.241 |
0.139 |
| 480 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.241 |
0.047 |
| 481 |
257321_at |
ATMG01130
|
ORF106F |
-0.240 |
0.012 |
| 482 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.240 |
-0.058 |
| 483 |
253386_at |
AT4G33030
|
SQD1, sulfoquinovosyldiacylglycerol 1 |
-0.239 |
0.004 |
| 484 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
-0.239 |
0.043 |
| 485 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
-0.239 |
0.050 |
| 486 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
-0.239 |
-0.027 |
| 487 |
247290_at |
AT5G64450
|
unknown |
-0.238 |
0.013 |
| 488 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.237 |
0.373 |
| 489 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.237 |
-0.081 |
| 490 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
-0.237 |
0.443 |
| 491 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.236 |
-0.008 |
| 492 |
246888_at |
AT5G26270
|
unknown |
-0.236 |
0.030 |
| 493 |
248329_at |
AT5G52780
|
unknown |
-0.236 |
-0.073 |
| 494 |
262291_at |
AT1G70790
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.235 |
-0.060 |
| 495 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.234 |
0.022 |
| 496 |
247851_at |
AT5G58070
|
ATTIL, TEMPERATURE-INDUCED LIPOCALIN, TIL, temperature-induced lipocalin |
-0.234 |
0.158 |
| 497 |
250925_at |
AT5G03370
|
[acylphosphatase family] |
-0.234 |
0.024 |
| 498 |
250127_at |
AT5G16380
|
unknown |
-0.234 |
-0.067 |
| 499 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
-0.234 |
0.097 |
| 500 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.233 |
-0.173 |
| 501 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
-0.233 |
0.167 |
| 502 |
256301_at |
AT1G69510
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.233 |
-0.027 |
| 503 |
246757_at |
AT5G27940
|
WPP3, WPP domain protein 3 |
-0.232 |
-0.027 |
| 504 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.232 |
0.015 |
| 505 |
261373_at |
AT1G53000
|
AtCKS, CKS, CMP-KDO synthetase, KDSB |
-0.231 |
0.034 |
| 506 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.231 |
0.018 |
| 507 |
257483_at |
AT1G49620
|
AtKRP7, ICK5, ICN6, KRP7, KIP-RELATED PROTEIN 7 |
-0.231 |
-0.177 |
| 508 |
254211_at |
AT4G23570
|
SGT1A |
-0.231 |
-0.055 |
| 509 |
263032_at |
AT1G23850
|
unknown |
-0.230 |
-0.007 |
| 510 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
-0.228 |
-0.001 |
| 511 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.228 |
-0.051 |
| 512 |
249177_at |
AT5G42850
|
[Thioredoxin superfamily protein] |
-0.227 |
0.039 |
| 513 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.227 |
-0.102 |
| 514 |
248485_at |
AT5G51050
|
APC2, ATP/phosphate carrier 2 |
-0.227 |
0.035 |
| 515 |
252973_s_at |
AT4G38740
|
ROC1, rotamase CYP 1 |
-0.226 |
-0.048 |
| 516 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
-0.225 |
-0.017 |
| 517 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
-0.224 |
-0.022 |
| 518 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.224 |
0.188 |
| 519 |
266673_at |
AT2G29630
|
PY, PYRIMIDINE REQUIRING, THIC, thiaminC |
-0.223 |
-0.034 |
| 520 |
262014_at |
AT1G35660
|
unknown |
-0.223 |
-0.066 |
| 521 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.223 |
-0.140 |
| 522 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
-0.223 |
-0.129 |
| 523 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
-0.223 |
-0.125 |
| 524 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
-0.223 |
0.059 |
| 525 |
267521_at |
AT2G30480
|
unknown |
-0.223 |
0.021 |
| 526 |
267405_at |
AT2G33740
|
CUTA |
-0.222 |
0.018 |
| 527 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.222 |
-0.127 |
| 528 |
255590_at |
AT4G01610
|
[Cysteine proteinases superfamily protein] |
-0.222 |
-0.024 |
| 529 |
261655_at |
AT1G01940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.222 |
-0.024 |
| 530 |
259666_at |
AT1G55310
|
At-SCL33, SC35-LIKE SPLICING FACTOR 33, ATSCL33, SC35-like splicing factor 33, SCL33, SC35-LIKE SPLICING FACTOR 33, SR33, SC35-like splicing factor 33 |
-0.222 |
-0.094 |
| 531 |
263649_at |
AT1G04380
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.222 |
0.039 |
| 532 |
264086_at |
AT2G31190
|
RUS2, ROOT UV-B SENSITIVE 2, WXR1, weak auxin response1 |
-0.221 |
-0.088 |
| 533 |
263881_at |
AT2G21820
|
unknown |
-0.221 |
0.074 |
| 534 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.220 |
-0.179 |
| 535 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
-0.220 |
0.425 |
| 536 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
-0.220 |
0.041 |
| 537 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
-0.220 |
-0.102 |
| 538 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.219 |
-0.070 |
| 539 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
-0.219 |
0.176 |
| 540 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
-0.219 |
0.061 |
| 541 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.219 |
-0.040 |
| 542 |
250074_at |
AT5G17310
|
AtUGP2, UDP-GLUCOSE PYROPHOSPHORYLASE 2, UGP2, UDP-glucose pyrophosphorylase 2 |
-0.219 |
-0.007 |
| 543 |
249272_at |
AT5G41820
|
ATRGTA2, RAB geranylgeranyl transferase alpha subunit 2, RGTA2, RAB geranylgeranyl transferase alpha subunit 2 |
-0.218 |
-0.006 |