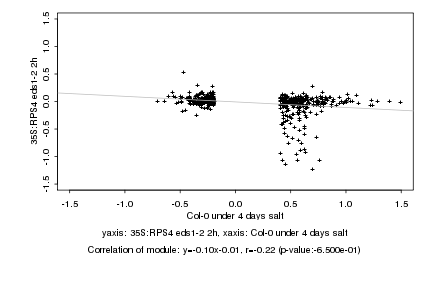
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 under 4 days salt |
Log2 signal ratio
35S:RPS4 eds1-2 2h |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.490 |
-0.016 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.389 |
0.009 |
| 3 |
251635_at |
AT3G57510
|
ADPG1 |
1.285 |
0.010 |
| 4 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.232 |
-0.063 |
| 5 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.226 |
0.029 |
| 6 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.215 |
-0.055 |
| 7 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.111 |
-0.034 |
| 8 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.088 |
0.111 |
| 9 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
1.057 |
0.011 |
| 10 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
1.041 |
0.004 |
| 11 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.014 |
0.063 |
| 12 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
1.006 |
0.145 |
| 13 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.003 |
-0.007 |
| 14 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.002 |
0.027 |
| 15 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.989 |
0.017 |
| 16 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.980 |
-0.006 |
| 17 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.973 |
-0.029 |
| 18 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.953 |
0.013 |
| 19 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.940 |
-0.035 |
| 20 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.935 |
0.087 |
| 21 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.906 |
-0.069 |
| 22 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.887 |
0.007 |
| 23 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.880 |
0.042 |
| 24 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.872 |
0.006 |
| 25 |
264580_at |
AT1G05340
|
unknown |
0.866 |
-0.080 |
| 26 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.864 |
-0.037 |
| 27 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.855 |
0.080 |
| 28 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.848 |
0.087 |
| 29 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.828 |
0.032 |
| 30 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.820 |
0.051 |
| 31 |
256114_at |
AT1G16850
|
unknown |
0.815 |
0.055 |
| 32 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.813 |
-0.018 |
| 33 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.813 |
0.084 |
| 34 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.806 |
0.053 |
| 35 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.804 |
-0.010 |
| 36 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.801 |
-0.006 |
| 37 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.795 |
-0.082 |
| 38 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.792 |
-0.052 |
| 39 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.791 |
-0.030 |
| 40 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.789 |
-0.007 |
| 41 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.786 |
0.090 |
| 42 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.784 |
0.005 |
| 43 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.783 |
0.046 |
| 44 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.780 |
0.173 |
| 45 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.776 |
-0.021 |
| 46 |
263475_at |
AT2G31945
|
unknown |
0.771 |
-0.172 |
| 47 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.769 |
0.064 |
| 48 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.765 |
0.092 |
| 49 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.763 |
-0.083 |
| 50 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.755 |
-1.067 |
| 51 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.751 |
-0.047 |
| 52 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.751 |
-0.009 |
| 53 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.750 |
0.017 |
| 54 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.742 |
-0.038 |
| 55 |
253859_at |
AT4G27657
|
unknown |
0.738 |
0.033 |
| 56 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.732 |
-0.225 |
| 57 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.732 |
0.017 |
| 58 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.732 |
-0.648 |
| 59 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.731 |
-0.009 |
| 60 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.729 |
0.083 |
| 61 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.727 |
-0.065 |
| 62 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.705 |
0.004 |
| 63 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.704 |
-0.040 |
| 64 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.699 |
-0.061 |
| 65 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.697 |
0.280 |
| 66 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.689 |
0.067 |
| 67 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.689 |
-1.226 |
| 68 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.684 |
0.052 |
| 69 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.676 |
-0.199 |
| 70 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.675 |
0.052 |
| 71 |
249377_at |
AT5G40690
|
unknown |
0.663 |
-0.027 |
| 72 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.655 |
-0.006 |
| 73 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.651 |
0.076 |
| 74 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.649 |
-0.091 |
| 75 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.646 |
0.033 |
| 76 |
261243_at |
AT1G20180
|
unknown |
0.644 |
-0.059 |
| 77 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.641 |
0.067 |
| 78 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.640 |
-0.061 |
| 79 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.640 |
0.093 |
| 80 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.638 |
-0.050 |
| 81 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.638 |
-0.276 |
| 82 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.638 |
0.040 |
| 83 |
260744_at |
AT1G15010
|
unknown |
0.635 |
-0.124 |
| 84 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.634 |
0.116 |
| 85 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.630 |
0.101 |
| 86 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.630 |
-0.253 |
| 87 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.629 |
-0.115 |
| 88 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.628 |
0.028 |
| 89 |
256442_at |
AT3G10930
|
unknown |
0.626 |
-0.919 |
| 90 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.622 |
0.025 |
| 91 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.621 |
-0.482 |
| 92 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.620 |
-0.182 |
| 93 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.618 |
-0.445 |
| 94 |
265276_at |
AT2G28400
|
unknown |
0.618 |
-0.591 |
| 95 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.616 |
0.028 |
| 96 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.616 |
-0.857 |
| 97 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.615 |
0.157 |
| 98 |
253796_at |
AT4G28460
|
unknown |
0.613 |
-0.086 |
| 99 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.613 |
-0.181 |
| 100 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.613 |
0.060 |
| 101 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.610 |
0.032 |
| 102 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.605 |
-0.050 |
| 103 |
251766_at |
AT3G55910
|
unknown |
0.603 |
0.001 |
| 104 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.602 |
-0.004 |
| 105 |
253643_at |
AT4G29780
|
unknown |
0.596 |
-0.749 |
| 106 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.594 |
-0.018 |
| 107 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.592 |
-0.031 |
| 108 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.592 |
0.048 |
| 109 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
0.590 |
0.025 |
| 110 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.590 |
0.081 |
| 111 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.589 |
0.042 |
| 112 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.587 |
0.047 |
| 113 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.586 |
0.095 |
| 114 |
260841_at |
AT1G29195
|
unknown |
0.583 |
-0.002 |
| 115 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.582 |
-0.876 |
| 116 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.582 |
0.095 |
| 117 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.582 |
-0.091 |
| 118 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.581 |
-0.042 |
| 119 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.579 |
-0.071 |
| 120 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.577 |
-0.151 |
| 121 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.577 |
-0.037 |
| 122 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.576 |
0.005 |
| 123 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.574 |
-0.035 |
| 124 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
0.573 |
0.043 |
| 125 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.573 |
-0.298 |
| 126 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.572 |
0.017 |
| 127 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.571 |
-0.192 |
| 128 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.571 |
-0.523 |
| 129 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.571 |
-0.329 |
| 130 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.570 |
0.058 |
| 131 |
254289_at |
AT4G22980
|
unknown |
0.570 |
-0.123 |
| 132 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.568 |
-0.050 |
| 133 |
253827_at |
AT4G28085
|
unknown |
0.567 |
-0.227 |
| 134 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.567 |
0.108 |
| 135 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.565 |
0.054 |
| 136 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.564 |
-0.707 |
| 137 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.564 |
0.054 |
| 138 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.560 |
-0.094 |
| 139 |
258209_at |
AT3G14060
|
unknown |
0.560 |
0.014 |
| 140 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.557 |
-0.198 |
| 141 |
263182_at |
AT1G05575
|
unknown |
0.557 |
-1.071 |
| 142 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.553 |
0.015 |
| 143 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
0.552 |
-0.031 |
| 144 |
267178_at |
AT2G37750
|
unknown |
0.551 |
0.029 |
| 145 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.545 |
-0.947 |
| 146 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.545 |
0.055 |
| 147 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.542 |
0.081 |
| 148 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.540 |
0.079 |
| 149 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.538 |
0.017 |
| 150 |
256407_at |
AT1G66570
|
ATSUC7, sucrose-proton symporter 7, SUC7, sucrose-proton symporter 7 |
0.537 |
0.071 |
| 151 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.536 |
-0.046 |
| 152 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.532 |
0.006 |
| 153 |
265184_at |
AT1G23710
|
unknown |
0.531 |
-0.470 |
| 154 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.530 |
0.022 |
| 155 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.529 |
0.025 |
| 156 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.528 |
0.030 |
| 157 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.527 |
0.018 |
| 158 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.527 |
0.039 |
| 159 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.527 |
-0.009 |
| 160 |
260522_x_at |
AT2G41730
|
unknown |
0.526 |
-0.059 |
| 161 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.524 |
-0.008 |
| 162 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.524 |
-0.060 |
| 163 |
248505_at |
AT5G50360
|
unknown |
0.523 |
-0.012 |
| 164 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.522 |
-0.050 |
| 165 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.522 |
0.013 |
| 166 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.522 |
-0.003 |
| 167 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.521 |
0.062 |
| 168 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.521 |
0.054 |
| 169 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.521 |
0.013 |
| 170 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.520 |
0.019 |
| 171 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.520 |
0.010 |
| 172 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.519 |
0.027 |
| 173 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.518 |
-0.026 |
| 174 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.518 |
-0.001 |
| 175 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.518 |
-0.098 |
| 176 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.517 |
-0.233 |
| 177 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.514 |
0.021 |
| 178 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.514 |
0.042 |
| 179 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
0.514 |
-0.004 |
| 180 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.513 |
-0.226 |
| 181 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.513 |
-0.660 |
| 182 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.512 |
-0.066 |
| 183 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.510 |
0.156 |
| 184 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.509 |
0.002 |
| 185 |
251293_at |
AT3G61930
|
unknown |
0.509 |
0.010 |
| 186 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.504 |
-0.298 |
| 187 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.502 |
-0.023 |
| 188 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.500 |
0.017 |
| 189 |
251019_at |
AT5G02420
|
unknown |
0.500 |
0.059 |
| 190 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.499 |
-0.036 |
| 191 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.498 |
0.046 |
| 192 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.498 |
0.001 |
| 193 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.495 |
0.014 |
| 194 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.492 |
0.012 |
| 195 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.492 |
-0.281 |
| 196 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.490 |
-0.390 |
| 197 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.489 |
0.017 |
| 198 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.488 |
0.023 |
| 199 |
260411_at |
AT1G69890
|
unknown |
0.487 |
-0.267 |
| 200 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.487 |
0.029 |
| 201 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.487 |
0.032 |
| 202 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.486 |
0.015 |
| 203 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.486 |
0.024 |
| 204 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.483 |
-0.029 |
| 205 |
253832_at |
AT4G27654
|
unknown |
0.481 |
-0.006 |
| 206 |
267226_at |
AT2G44010
|
unknown |
0.481 |
-0.018 |
| 207 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.481 |
-0.022 |
| 208 |
253830_at |
AT4G27652
|
unknown |
0.481 |
-0.001 |
| 209 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.481 |
-0.001 |
| 210 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.480 |
-0.749 |
| 211 |
246183_at |
AT5G20940
|
[Glycosyl hydrolase family protein] |
0.479 |
0.018 |
| 212 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.478 |
-0.006 |
| 213 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.477 |
-0.014 |
| 214 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.477 |
-0.151 |
| 215 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.477 |
-0.019 |
| 216 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.471 |
-0.003 |
| 217 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.471 |
-0.329 |
| 218 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.469 |
-0.039 |
| 219 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.468 |
-0.000 |
| 220 |
249454_at |
AT5G39520
|
unknown |
0.468 |
0.109 |
| 221 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.468 |
-0.268 |
| 222 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.466 |
0.013 |
| 223 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.466 |
-0.622 |
| 224 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.465 |
-0.121 |
| 225 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.465 |
0.076 |
| 226 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.463 |
-0.035 |
| 227 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.462 |
-0.007 |
| 228 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.461 |
-0.087 |
| 229 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.459 |
-0.166 |
| 230 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
0.459 |
-0.080 |
| 231 |
265913_at |
AT2G25625
|
unknown |
0.458 |
0.063 |
| 232 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.458 |
0.099 |
| 233 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.458 |
0.019 |
| 234 |
265059_at |
AT1G52080
|
AR791 |
0.457 |
0.017 |
| 235 |
252400_at |
AT3G48020
|
unknown |
0.456 |
0.024 |
| 236 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.456 |
0.013 |
| 237 |
259856_at |
AT1G68440
|
unknown |
0.456 |
-0.013 |
| 238 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.455 |
-0.012 |
| 239 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.455 |
0.095 |
| 240 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.454 |
0.081 |
| 241 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.454 |
0.067 |
| 242 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.452 |
0.117 |
| 243 |
257925_at |
AT3G23170
|
unknown |
0.452 |
-0.099 |
| 244 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.451 |
-0.347 |
| 245 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.450 |
-0.021 |
| 246 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.450 |
0.067 |
| 247 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.446 |
0.090 |
| 248 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.446 |
-1.133 |
| 249 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.445 |
-0.238 |
| 250 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.445 |
0.071 |
| 251 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.444 |
0.036 |
| 252 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.442 |
-0.009 |
| 253 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.441 |
-0.485 |
| 254 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.440 |
-0.025 |
| 255 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.439 |
0.034 |
| 256 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.438 |
0.111 |
| 257 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.438 |
-0.054 |
| 258 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.438 |
-0.082 |
| 259 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.437 |
-0.577 |
| 260 |
262211_at |
AT1G74930
|
ORA47 |
0.432 |
-0.093 |
| 261 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.432 |
0.057 |
| 262 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.432 |
0.009 |
| 263 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.432 |
-0.025 |
| 264 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.431 |
-0.051 |
| 265 |
247835_at |
AT5G57910
|
unknown |
0.430 |
-0.251 |
| 266 |
247488_at |
AT5G61820
|
unknown |
0.430 |
0.018 |
| 267 |
256861_at |
AT3G23920
|
AtBAM1, BAM1, beta-amylase 1, BMY7, BETA-AMYLASE 7, TR-BAMY |
0.429 |
0.025 |
| 268 |
255733_at |
AT1G25400
|
unknown |
0.428 |
-0.302 |
| 269 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.428 |
-0.061 |
| 270 |
247913_at |
AT5G57510
|
unknown |
0.427 |
-0.063 |
| 271 |
259979_at |
AT1G76600
|
unknown |
0.426 |
-0.377 |
| 272 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.426 |
0.023 |
| 273 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.425 |
-0.024 |
| 274 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.425 |
-1.065 |
| 275 |
247708_at |
AT5G59550
|
unknown |
0.424 |
-0.410 |
| 276 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.423 |
-0.043 |
| 277 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.423 |
-0.059 |
| 278 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.420 |
0.018 |
| 279 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.420 |
0.081 |
| 280 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.420 |
0.088 |
| 281 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.420 |
0.065 |
| 282 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.419 |
-0.197 |
| 283 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.419 |
0.058 |
| 284 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.418 |
0.118 |
| 285 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.418 |
0.129 |
| 286 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.417 |
0.012 |
| 287 |
263566_at |
AT2G15340
|
[glycine-rich protein] |
0.417 |
-0.041 |
| 288 |
256014_at |
AT1G19200
|
unknown |
0.417 |
0.013 |
| 289 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.416 |
0.030 |
| 290 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.412 |
0.071 |
| 291 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.408 |
-0.406 |
| 292 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.406 |
0.075 |
| 293 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.406 |
0.057 |
| 294 |
264660_at |
AT1G09940
|
HEMA2 |
0.406 |
-0.051 |
| 295 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.406 |
0.012 |
| 296 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.405 |
0.031 |
| 297 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.405 |
0.061 |
| 298 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.403 |
-0.942 |
| 299 |
267036_at |
AT2G38465
|
unknown |
0.403 |
-0.056 |
| 300 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.402 |
-0.117 |
| 301 |
250327_at |
AT5G12050
|
unknown |
-0.709 |
0.012 |
| 302 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.643 |
0.012 |
| 303 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.606 |
0.095 |
| 304 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.576 |
0.179 |
| 305 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.567 |
0.103 |
| 306 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.544 |
0.080 |
| 307 |
249932_at |
AT5G22390
|
unknown |
-0.539 |
-0.022 |
| 308 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.517 |
-0.010 |
| 309 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.502 |
0.071 |
| 310 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.500 |
0.106 |
| 311 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.493 |
-0.016 |
| 312 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.489 |
0.016 |
| 313 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.483 |
0.077 |
| 314 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.482 |
-0.181 |
| 315 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.472 |
0.542 |
| 316 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.460 |
-0.161 |
| 317 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.445 |
0.061 |
| 318 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.442 |
0.064 |
| 319 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.432 |
0.022 |
| 320 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.424 |
-0.021 |
| 321 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.423 |
0.175 |
| 322 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.422 |
0.053 |
| 323 |
258196_at |
AT3G13980
|
unknown |
-0.420 |
0.081 |
| 324 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.416 |
0.109 |
| 325 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.415 |
0.050 |
| 326 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.411 |
0.001 |
| 327 |
248937_at |
AT5G45770
|
AtRLP55, receptor like protein 55, RLP55, receptor like protein 55 |
-0.411 |
-0.031 |
| 328 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.408 |
-0.047 |
| 329 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.404 |
0.046 |
| 330 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.395 |
0.005 |
| 331 |
262162_at |
AT1G78020
|
unknown |
-0.382 |
0.015 |
| 332 |
249332_at |
AT5G40980
|
unknown |
-0.382 |
-0.041 |
| 333 |
251058_at |
AT5G01790
|
unknown |
-0.382 |
0.049 |
| 334 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.379 |
0.049 |
| 335 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.378 |
-0.029 |
| 336 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.377 |
0.006 |
| 337 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.373 |
0.028 |
| 338 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.369 |
0.039 |
| 339 |
250936_at |
AT5G03120
|
unknown |
-0.368 |
-0.024 |
| 340 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.367 |
0.055 |
| 341 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.367 |
-0.006 |
| 342 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.365 |
0.003 |
| 343 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.364 |
0.100 |
| 344 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.364 |
-0.029 |
| 345 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.360 |
-0.048 |
| 346 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.358 |
0.116 |
| 347 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.357 |
-0.242 |
| 348 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.354 |
0.117 |
| 349 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.353 |
-0.005 |
| 350 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.352 |
-0.022 |
| 351 |
253243_at |
AT4G34560
|
unknown |
-0.349 |
0.303 |
| 352 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.347 |
0.110 |
| 353 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.345 |
0.015 |
| 354 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.341 |
0.125 |
| 355 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.341 |
0.050 |
| 356 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.341 |
0.007 |
| 357 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.339 |
0.076 |
| 358 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.338 |
0.046 |
| 359 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.336 |
-0.000 |
| 360 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.333 |
-0.055 |
| 361 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.332 |
-0.050 |
| 362 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.331 |
0.004 |
| 363 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.331 |
0.144 |
| 364 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.329 |
-0.054 |
| 365 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.328 |
0.013 |
| 366 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.327 |
0.035 |
| 367 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.327 |
0.005 |
| 368 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.326 |
0.130 |
| 369 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
-0.325 |
0.013 |
| 370 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.324 |
0.048 |
| 371 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.324 |
0.021 |
| 372 |
260363_at |
AT1G70550
|
unknown |
-0.321 |
0.014 |
| 373 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.320 |
-0.029 |
| 374 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.320 |
0.157 |
| 375 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.319 |
-0.011 |
| 376 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.317 |
0.029 |
| 377 |
247878_at |
AT5G57760
|
unknown |
-0.317 |
-0.001 |
| 378 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.316 |
0.059 |
| 379 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.316 |
0.166 |
| 380 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.315 |
-0.049 |
| 381 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.315 |
0.020 |
| 382 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
-0.313 |
0.069 |
| 383 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.312 |
-0.010 |
| 384 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.312 |
0.003 |
| 385 |
261223_at |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
-0.311 |
0.048 |
| 386 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.311 |
-0.091 |
| 387 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.310 |
-0.008 |
| 388 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.309 |
0.012 |
| 389 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.305 |
0.033 |
| 390 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.304 |
-0.051 |
| 391 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.304 |
0.059 |
| 392 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.303 |
-0.020 |
| 393 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.301 |
0.018 |
| 394 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.300 |
0.172 |
| 395 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.300 |
-0.010 |
| 396 |
261247_at |
AT1G20070
|
unknown |
-0.300 |
0.065 |
| 397 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.298 |
0.040 |
| 398 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.297 |
0.046 |
| 399 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
-0.296 |
0.128 |
| 400 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.291 |
-0.006 |
| 401 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.291 |
-0.013 |
| 402 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.289 |
0.079 |
| 403 |
252119_at |
AT3G51030
|
ATTRX H1, ARABIDOPSIS THALIANA THIOREDOXIN H-TYPE 1, ATTRX1, thioredoxin H-type 1, TRX1, thioredoxin H-type 1 |
-0.289 |
-0.110 |
| 404 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.289 |
0.013 |
| 405 |
263535_at |
AT2G24970
|
unknown |
-0.288 |
0.021 |
| 406 |
260112_at |
AT1G63310
|
unknown |
-0.288 |
0.048 |
| 407 |
258421_at |
AT3G16690
|
AtSWEET16, SWEET16 |
-0.287 |
-0.023 |
| 408 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.287 |
0.010 |
| 409 |
247101_at |
AT5G66530
|
[Galactose mutarotase-like superfamily protein] |
-0.284 |
-0.013 |
| 410 |
258232_at |
AT3G27750
|
EMB3123, EMBRYO DEFECTIVE 3123, THA8, Thylakoid Assembly 8 |
-0.284 |
0.039 |
| 411 |
248167_at |
AT5G54530
|
unknown |
-0.283 |
0.000 |
| 412 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.283 |
-0.016 |
| 413 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
-0.282 |
0.024 |
| 414 |
248028_at |
AT5G55620
|
unknown |
-0.282 |
0.012 |
| 415 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.281 |
0.001 |
| 416 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.281 |
0.079 |
| 417 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.280 |
0.044 |
| 418 |
260460_at |
AT1G68230
|
[Reticulon family protein] |
-0.279 |
0.005 |
| 419 |
264987_at |
AT1G27030
|
unknown |
-0.277 |
-0.015 |
| 420 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.277 |
0.083 |
| 421 |
250189_at |
AT5G14410
|
unknown |
-0.277 |
0.040 |
| 422 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.276 |
-0.013 |
| 423 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.276 |
0.041 |
| 424 |
252025_at |
AT3G52900
|
unknown |
-0.274 |
0.079 |
| 425 |
245426_at |
AT4G17540
|
unknown |
-0.272 |
0.023 |
| 426 |
249162_at |
AT5G42765
|
unknown |
-0.271 |
0.004 |
| 427 |
264293_at |
AT1G78770
|
APC6, anaphase promoting complex 6 |
-0.271 |
0.042 |
| 428 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.271 |
0.007 |
| 429 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.270 |
-0.008 |
| 430 |
262539_at |
AT1G17200
|
[Uncharacterised protein family (UPF0497)] |
-0.269 |
0.010 |
| 431 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.268 |
0.004 |
| 432 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.268 |
0.035 |
| 433 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.265 |
0.014 |
| 434 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
-0.265 |
0.010 |
| 435 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.265 |
0.025 |
| 436 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.264 |
0.017 |
| 437 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
-0.263 |
0.080 |
| 438 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.263 |
0.093 |
| 439 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.263 |
0.051 |
| 440 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.263 |
0.104 |
| 441 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.262 |
0.027 |
| 442 |
260332_at |
AT1G70470
|
unknown |
-0.262 |
0.036 |
| 443 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.262 |
0.045 |
| 444 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.261 |
0.018 |
| 445 |
257134_at |
AT3G12870
|
unknown |
-0.261 |
0.015 |
| 446 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.261 |
0.012 |
| 447 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.261 |
0.006 |
| 448 |
250734_at |
AT5G06270
|
unknown |
-0.260 |
-0.033 |
| 449 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.260 |
0.035 |
| 450 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.259 |
0.074 |
| 451 |
259996_at |
AT1G67910
|
unknown |
-0.258 |
0.140 |
| 452 |
246746_at |
AT5G27820
|
[Ribosomal L18p/L5e family protein] |
-0.258 |
-0.010 |
| 453 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.257 |
0.070 |
| 454 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
-0.257 |
0.063 |
| 455 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.256 |
0.071 |
| 456 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.256 |
0.089 |
| 457 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.256 |
0.028 |
| 458 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.256 |
-0.119 |
| 459 |
248493_at |
AT5G51100
|
FSD2, Fe superoxide dismutase 2 |
-0.255 |
0.008 |
| 460 |
267636_at |
AT2G42110
|
unknown |
-0.254 |
0.020 |
| 461 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.254 |
-0.091 |
| 462 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.252 |
0.116 |
| 463 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.252 |
0.081 |
| 464 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.251 |
-0.069 |
| 465 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.250 |
0.012 |
| 466 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.246 |
0.042 |
| 467 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.244 |
0.042 |
| 468 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.244 |
0.011 |
| 469 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.243 |
0.012 |
| 470 |
263442_at |
AT2G28605
|
[Photosystem II reaction center PsbP family protein] |
-0.243 |
-0.016 |
| 471 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.242 |
0.018 |
| 472 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.242 |
0.056 |
| 473 |
261388_at |
AT1G05385
|
LPA19, LOW PSII ACCUMULATION 19, Psb27-H1 |
-0.242 |
-0.018 |
| 474 |
249915_at |
AT5G22870
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.240 |
0.066 |
| 475 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.240 |
0.024 |
| 476 |
251778_at |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
-0.240 |
0.050 |
| 477 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.240 |
0.066 |
| 478 |
257974_at |
AT3G20820
|
[Leucine-rich repeat (LRR) family protein] |
-0.240 |
0.041 |
| 479 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.239 |
-0.064 |
| 480 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.239 |
0.020 |
| 481 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.238 |
0.008 |
| 482 |
264385_at |
AT1G12020
|
unknown |
-0.238 |
0.052 |
| 483 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.238 |
0.047 |
| 484 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.238 |
0.020 |
| 485 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.238 |
0.069 |
| 486 |
261577_at |
AT1G01080
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.238 |
0.012 |
| 487 |
266521_at |
AT2G24020
|
[Uncharacterised BCR, YbaB family COG0718] |
-0.237 |
-0.002 |
| 488 |
253045_at |
AT4G37445
|
unknown |
-0.236 |
0.035 |
| 489 |
256447_at |
AT1G33440
|
AtNPF4.4, NPF4.4, NRT1/ PTR family 4.4 |
-0.236 |
0.024 |
| 490 |
267367_at |
AT2G44210
|
unknown |
-0.236 |
0.022 |
| 491 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.235 |
0.022 |
| 492 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
-0.235 |
-0.005 |
| 493 |
255558_at |
AT4G01900
|
GLB1, GLNB1 homolog, PII |
-0.235 |
0.001 |
| 494 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.235 |
-0.013 |
| 495 |
263002_at |
AT1G54200
|
unknown |
-0.234 |
0.096 |
| 496 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.234 |
-0.003 |
| 497 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
-0.234 |
0.040 |
| 498 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.234 |
0.006 |
| 499 |
251788_at |
AT3G55420
|
unknown |
-0.231 |
0.046 |
| 500 |
247457_at |
AT5G62170
|
TRM25, TON1 Recruiting Motif 25 |
-0.231 |
-0.034 |
| 501 |
245309_at |
AT4G15140
|
unknown |
-0.230 |
-0.027 |
| 502 |
248623_at |
AT5G49170
|
unknown |
-0.230 |
0.096 |
| 503 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.230 |
0.024 |
| 504 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.230 |
-0.130 |
| 505 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.230 |
0.015 |
| 506 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.229 |
0.079 |
| 507 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.229 |
-0.005 |
| 508 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.229 |
0.052 |
| 509 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.229 |
-0.006 |
| 510 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.228 |
0.169 |
| 511 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.228 |
-0.016 |
| 512 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.227 |
0.151 |
| 513 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
-0.225 |
0.068 |
| 514 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.225 |
0.119 |
| 515 |
252001_at |
AT3G52750
|
FTSZ2-2 |
-0.225 |
0.020 |
| 516 |
253740_at |
AT4G28706
|
[pfkB-like carbohydrate kinase family protein] |
-0.225 |
0.032 |
| 517 |
260673_at |
AT1G19330
|
unknown |
-0.225 |
-0.044 |
| 518 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.225 |
0.073 |
| 519 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.224 |
0.080 |
| 520 |
253475_at |
AT4G32290
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.224 |
0.127 |
| 521 |
261178_at |
AT1G04760
|
ATVAMP726, vesicle-associated membrane protein 726, VAMP726, vesicle-associated membrane protein 726 |
-0.224 |
0.045 |
| 522 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.224 |
-0.007 |
| 523 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.223 |
0.006 |
| 524 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.222 |
0.035 |
| 525 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.221 |
-0.003 |
| 526 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.221 |
-0.014 |
| 527 |
266704_at |
AT2G19940
|
[oxidoreductases, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor] |
-0.221 |
0.001 |
| 528 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.221 |
0.032 |
| 529 |
260441_at |
AT1G68260
|
ALT3, acyl-lipid thioesterase 3 |
-0.221 |
-0.002 |
| 530 |
257237_at |
AT3G14890
|
[phosphoesterase] |
-0.221 |
0.006 |
| 531 |
256728_at |
AT3G25660
|
[Amidase family protein] |
-0.221 |
0.038 |
| 532 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.220 |
0.052 |
| 533 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
-0.219 |
0.004 |
| 534 |
266662_at |
AT2G25830
|
[YebC-related] |
-0.219 |
0.015 |
| 535 |
253920_at |
AT4G27230
|
HTA2, histone H2A 2 |
-0.218 |
0.020 |
| 536 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.218 |
0.123 |
| 537 |
262693_at |
AT1G62780
|
unknown |
-0.218 |
-0.009 |
| 538 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.218 |
0.037 |
| 539 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
-0.217 |
0.065 |
| 540 |
267356_at |
AT2G39930
|
ATISA1, ARABIDOPSIS THALIANA ISOAMYLASE 1, ISA1, isoamylase 1 |
-0.217 |
-0.001 |
| 541 |
249986_at |
AT5G18460
|
unknown |
-0.217 |
0.040 |
| 542 |
250696_at |
AT5G06790
|
unknown |
-0.217 |
0.054 |
| 543 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.217 |
0.023 |
| 544 |
266635_at |
AT2G35470
|
unknown |
-0.216 |
0.040 |
| 545 |
247153_at |
AT5G65700
|
BAM1, BARELY ANY MERISTEM 1 |
-0.216 |
-0.029 |
| 546 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
-0.215 |
0.035 |
| 547 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.215 |
0.047 |
| 548 |
259181_at |
AT3G01690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.215 |
0.011 |
| 549 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.215 |
0.041 |
| 550 |
262811_at |
AT1G11700
|
unknown |
-0.214 |
0.082 |
| 551 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.214 |
0.010 |
| 552 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.214 |
0.033 |
| 553 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.214 |
0.028 |
| 554 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.214 |
0.167 |
| 555 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.213 |
0.032 |
| 556 |
259288_at |
AT3G11500
|
[Small nuclear ribonucleoprotein family protein] |
-0.212 |
0.019 |
| 557 |
252501_at |
AT3G46880
|
unknown |
-0.212 |
0.289 |
| 558 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.212 |
0.029 |
| 559 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.212 |
-0.007 |
| 560 |
253607_at |
AT4G30330
|
[Small nuclear ribonucleoprotein family protein] |
-0.212 |
-0.052 |
| 561 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.211 |
-0.022 |
| 562 |
252274_at |
AT3G49680
|
ATBCAT-3, BCAT3, branched-chain aminotransferase 3 |
-0.211 |
0.061 |
| 563 |
259674_at |
AT1G77700
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.211 |
0.013 |
| 564 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.210 |
0.159 |
| 565 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.210 |
0.012 |
| 566 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.209 |
0.085 |
| 567 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.209 |
-0.010 |
| 568 |
249211_at |
AT5G42680
|
unknown |
-0.209 |
0.134 |
| 569 |
266882_at |
AT2G44670
|
unknown |
-0.208 |
0.012 |
| 570 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.208 |
0.050 |
| 571 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
-0.207 |
0.038 |
| 572 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.207 |
0.026 |
| 573 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
-0.207 |
0.021 |
| 574 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.206 |
0.073 |
| 575 |
251415_at |
AT3G60380
|
unknown |
-0.206 |
-0.022 |
| 576 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
-0.206 |
0.059 |
| 577 |
246547_at |
AT5G14970
|
unknown |
-0.205 |
-0.008 |
| 578 |
264352_at |
AT1G03270
|
unknown |
-0.204 |
0.052 |
| 579 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.204 |
0.043 |
| 580 |
266570_at |
AT2G24090
|
PRPL35, plastid ribosomal protein L35 |
-0.204 |
-0.005 |
| 581 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.204 |
-0.037 |
| 582 |
265311_at |
AT2G20250
|
unknown |
-0.204 |
0.102 |
| 583 |
250198_at |
AT5G14100
|
ABCI11, ATP-binding cassette I11, ATNAP14, ARABIDOPSIS THALIANANON-INTRINSIC ABC PROTEIN 14, NAP14, non-intrinsic ABC protein 14 |
-0.203 |
-0.023 |
| 584 |
251083_at |
AT5G01590
|
TIC56, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 56 |
-0.203 |
0.052 |
| 585 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.203 |
-0.052 |
| 586 |
251736_at |
AT3G56130
|
[biotin/lipoyl attachment domain-containing protein] |
-0.203 |
0.025 |
| 587 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.203 |
0.080 |
| 588 |
249330_at |
AT5G40970
|
unknown |
-0.202 |
-0.025 |
| 589 |
246202_at |
AT4G37040
|
MAP1D, methionine aminopeptidase 1D |
-0.201 |
0.011 |
| 590 |
251598_at |
AT3G57600
|
[Integrase-type DNA-binding superfamily protein] |
-0.201 |
-0.001 |
| 591 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
-0.201 |
0.168 |
| 592 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.201 |
-0.019 |
| 593 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.201 |
0.004 |
| 594 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.200 |
0.013 |
| 595 |
259857_at |
AT1G68430
|
unknown |
-0.200 |
0.065 |
| 596 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.200 |
0.009 |
| 597 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.199 |
-0.058 |
| 598 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.199 |
-0.027 |
| 599 |
261460_at |
AT1G07880
|
ATMPK13 |
-0.199 |
0.139 |
| 600 |
261346_at |
AT1G79720
|
[Eukaryotic aspartyl protease family protein] |
-0.198 |
0.014 |