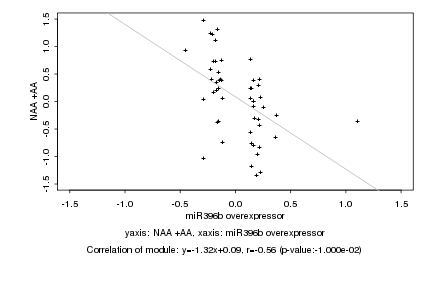
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
miR396b overexpressor |
Log2 signal ratio
NAA +AA |
| 1 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.099 |
-0.358 |
| 2 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.369 |
-0.254 |
| 3 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.355 |
-0.646 |
| 4 |
259765_at |
AT1G64370
|
unknown |
0.248 |
-0.095 |
| 5 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
0.226 |
0.078 |
| 6 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
0.225 |
-1.273 |
| 7 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
0.216 |
-0.427 |
| 8 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.215 |
0.409 |
| 9 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.211 |
-0.827 |
| 10 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.204 |
0.295 |
| 11 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.203 |
-0.320 |
| 12 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.192 |
-0.963 |
| 13 |
262396_at |
AT1G49470
|
unknown |
0.187 |
-1.333 |
| 14 |
248004_at |
AT5G56230
|
PRA1.G2, prenylated RAB acceptor 1.G2 |
0.164 |
-0.308 |
| 15 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
0.161 |
-0.087 |
| 16 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.158 |
0.012 |
| 17 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.158 |
0.398 |
| 18 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.157 |
-0.786 |
| 19 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.155 |
0.006 |
| 20 |
255646_at |
AT4G00890
|
[proline-rich family protein] |
0.138 |
0.250 |
| 21 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
0.138 |
-0.760 |
| 22 |
247874_at |
AT5G57710
|
SMAX1, SUPPRESSOR OF MAX2 1 |
0.137 |
-1.172 |
| 23 |
259546_at |
AT1G35350
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.131 |
-0.554 |
| 24 |
261599_at |
AT1G49700
|
unknown |
0.129 |
0.063 |
| 25 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.129 |
0.768 |
| 26 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
0.128 |
0.239 |
| 27 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
-0.455 |
0.943 |
| 28 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.297 |
-1.033 |
| 29 |
248335_at |
AT5G52450
|
[MATE efflux family protein] |
-0.295 |
0.053 |
| 30 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.295 |
1.491 |
| 31 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.230 |
1.255 |
| 32 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.228 |
0.590 |
| 33 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.221 |
0.405 |
| 34 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
-0.208 |
1.224 |
| 35 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.205 |
0.744 |
| 36 |
266825_at |
AT2G22890
|
[Kua-ubiquitin conjugating enzyme hybrid localisation domain] |
-0.199 |
0.176 |
| 37 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.189 |
0.741 |
| 38 |
260563_at |
AT2G43840
|
UGT74F1, UDP-glycosyltransferase 74 F1 |
-0.185 |
1.127 |
| 39 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
-0.174 |
0.203 |
| 40 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
-0.173 |
0.347 |
| 41 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
-0.167 |
-0.380 |
| 42 |
250389_at |
AT5G11320
|
AtYUC4, YUC4, YUCCA4 |
-0.163 |
1.319 |
| 43 |
262197_at |
AT1G53910
|
RAP2.12, related to AP2 12 |
-0.161 |
-0.357 |
| 44 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
-0.160 |
0.532 |
| 45 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
-0.159 |
0.237 |
| 46 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.149 |
0.395 |
| 47 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.143 |
0.410 |
| 48 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
-0.132 |
0.748 |
| 49 |
259122_at |
AT3G02210
|
COBL1, COBRA-like protein 1 precursor |
-0.130 |
0.393 |
| 50 |
257923_at |
AT3G23160
|
unknown |
-0.124 |
-0.736 |
| 51 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.123 |
0.060 |