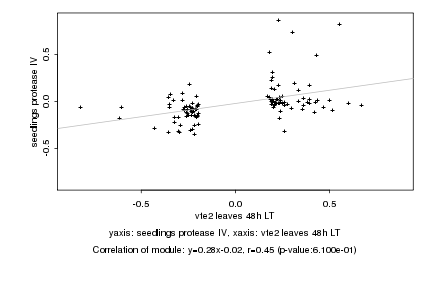
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
vte2 leaves 48h LT |
Log2 signal ratio
seedlings protease IV |
| 1 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.664 |
-0.039 |
| 2 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.596 |
-0.012 |
| 3 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.551 |
0.821 |
| 4 |
252938_at |
AT4G39190
|
unknown |
0.515 |
-0.092 |
| 5 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.499 |
0.019 |
| 6 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
0.467 |
-0.055 |
| 7 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
0.431 |
0.018 |
| 8 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.430 |
0.497 |
| 9 |
250211_at |
AT5G13880
|
unknown |
0.424 |
-0.007 |
| 10 |
248777_at |
AT5G47920
|
unknown |
0.418 |
-0.116 |
| 11 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.393 |
0.029 |
| 12 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.391 |
-0.021 |
| 13 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.390 |
0.180 |
| 14 |
260389_at |
AT1G74055
|
unknown |
0.382 |
-0.008 |
| 15 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.359 |
0.041 |
| 16 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.356 |
-0.041 |
| 17 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.351 |
-0.081 |
| 18 |
264635_at |
AT1G65500
|
unknown |
0.333 |
0.128 |
| 19 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.332 |
0.010 |
| 20 |
252289_at |
AT3G49130
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
0.311 |
0.193 |
| 21 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
0.300 |
0.737 |
| 22 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.296 |
-0.064 |
| 23 |
260277_at |
AT1G80520
|
[Sterile alpha motif (SAM) domain-containing protein] |
0.276 |
-0.028 |
| 24 |
247800_at |
AT5G58570
|
unknown |
0.258 |
-0.313 |
| 25 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.257 |
-0.037 |
| 26 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.256 |
-0.008 |
| 27 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.249 |
-0.018 |
| 28 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.245 |
0.063 |
| 29 |
251499_at |
AT3G59100
|
ATGSL11, glucan synthase-like 11, gsl11, GSL11, glucan synthase-like 11 |
0.239 |
0.013 |
| 30 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.236 |
-0.003 |
| 31 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.234 |
-0.101 |
| 32 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.232 |
-0.178 |
| 33 |
249438_at |
AT5G40010
|
AATP1, AAA-ATPase 1, ASD, ATPase-in-Seed-Development |
0.232 |
-0.013 |
| 34 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.232 |
0.045 |
| 35 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.226 |
0.874 |
| 36 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.225 |
0.172 |
| 37 |
249574_at |
AT5G37660
|
PDLP7, plasmodesmata-located protein 7 |
0.225 |
-0.013 |
| 38 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.221 |
0.024 |
| 39 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.214 |
0.031 |
| 40 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
0.213 |
-0.012 |
| 41 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.209 |
-0.005 |
| 42 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
0.206 |
-0.041 |
| 43 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.206 |
-0.026 |
| 44 |
257697_at |
AT3G12700
|
NANA, NANA |
0.205 |
0.136 |
| 45 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.201 |
-0.058 |
| 46 |
267345_at |
AT2G44240
|
unknown |
0.199 |
0.002 |
| 47 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.196 |
0.258 |
| 48 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
0.195 |
-0.001 |
| 49 |
267226_at |
AT2G44010
|
unknown |
0.194 |
0.317 |
| 50 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.192 |
0.029 |
| 51 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
0.191 |
0.232 |
| 52 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.189 |
0.142 |
| 53 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.188 |
0.008 |
| 54 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.187 |
-0.026 |
| 55 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
0.182 |
0.028 |
| 56 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
0.180 |
0.043 |
| 57 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.176 |
0.526 |
| 58 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.170 |
0.062 |
| 59 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.823 |
-0.054 |
| 60 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.616 |
-0.174 |
| 61 |
266938_at |
AT2G18950
|
ATHPT, HOMOGENTISATE PHYTYLTRANSFERASE, HPT1, homogentisate phytyltransferase 1, TPT1, VTE2, VITAMIN E 2 |
-0.605 |
-0.060 |
| 62 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.432 |
-0.281 |
| 63 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.357 |
0.050 |
| 64 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.356 |
-0.320 |
| 65 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.353 |
-0.031 |
| 66 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.351 |
-0.054 |
| 67 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.349 |
0.081 |
| 68 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.333 |
0.015 |
| 69 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
-0.327 |
-0.166 |
| 70 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.327 |
-0.214 |
| 71 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.307 |
-0.314 |
| 72 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.304 |
-0.161 |
| 73 |
253305_at |
AT4G33666
|
unknown |
-0.300 |
-0.326 |
| 74 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.296 |
-0.254 |
| 75 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.283 |
0.092 |
| 76 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.281 |
0.014 |
| 77 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.280 |
-0.084 |
| 78 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.275 |
-0.089 |
| 79 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.271 |
-0.055 |
| 80 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.265 |
-0.109 |
| 81 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.262 |
-0.050 |
| 82 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.260 |
-0.152 |
| 83 |
262751_at |
AT1G16310
|
[Cation efflux family protein] |
-0.259 |
-0.081 |
| 84 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.257 |
-0.126 |
| 85 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.253 |
-0.143 |
| 86 |
258225_at |
AT3G15630
|
unknown |
-0.249 |
0.183 |
| 87 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.245 |
-0.047 |
| 88 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.243 |
-0.060 |
| 89 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.243 |
-0.303 |
| 90 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.240 |
-0.078 |
| 91 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.238 |
-0.105 |
| 92 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.235 |
-0.143 |
| 93 |
259366_at |
AT1G13280
|
AOC4, allene oxide cyclase 4 |
-0.233 |
-0.073 |
| 94 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.233 |
-0.109 |
| 95 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.232 |
-0.108 |
| 96 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.232 |
-0.298 |
| 97 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.231 |
-0.016 |
| 98 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.228 |
-0.072 |
| 99 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.227 |
-0.106 |
| 100 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.226 |
-0.080 |
| 101 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.222 |
-0.351 |
| 102 |
259373_at |
AT1G69160
|
unknown |
-0.220 |
-0.143 |
| 103 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.218 |
-0.254 |
| 104 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.215 |
-0.159 |
| 105 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.213 |
-0.076 |
| 106 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
-0.211 |
-0.170 |
| 107 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.211 |
0.058 |
| 108 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.209 |
-0.080 |
| 109 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.206 |
-0.145 |
| 110 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.206 |
-0.151 |
| 111 |
267544_at |
AT2G32720
|
ATCB5-B, ARABIDOPSIS CYTOCHROME B5 ISOFORM B, B5 #4, CB5-B, cytochrome B5 isoform B, CYTB5-D |
-0.206 |
-0.051 |
| 112 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.202 |
-0.041 |
| 113 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.201 |
-0.238 |
| 114 |
258196_at |
AT3G13980
|
unknown |
-0.199 |
-0.122 |
| 115 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.197 |
-0.031 |