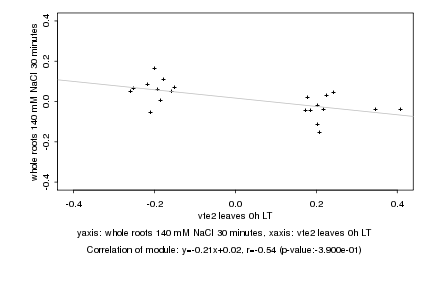
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
vte2 leaves 0h LT |
Log2 signal ratio
whole roots 140 mM NaCl 30 minutes |
| 1 |
264958_at |
AT1G76960
|
unknown |
0.407 |
-0.039 |
| 2 |
244934_at |
ATCG01080
|
NDHG |
0.344 |
-0.038 |
| 3 |
260217_at |
AT1G74600
|
OTP87, organelle transcript processing 87 |
0.240 |
0.045 |
| 4 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.225 |
0.032 |
| 5 |
244933_at |
ATCG01070
|
NDHE |
0.217 |
-0.038 |
| 6 |
263283_at |
AT2G36090
|
[F-box family protein] |
0.206 |
-0.153 |
| 7 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
0.201 |
-0.111 |
| 8 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.201 |
-0.019 |
| 9 |
252090_at |
AT3G52130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.185 |
-0.043 |
| 10 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.176 |
0.023 |
| 11 |
244997_at |
ATCG00170
|
RPOC2 |
0.173 |
-0.043 |
| 12 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
-0.260 |
0.053 |
| 13 |
255824_at |
AT2G40530
|
unknown |
-0.252 |
0.065 |
| 14 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.218 |
0.089 |
| 15 |
253809_at |
AT4G28320
|
AtMAN5, MAN5, endo-beta-mannase 5 |
-0.210 |
-0.050 |
| 16 |
245755_at |
AT1G35210
|
unknown |
-0.202 |
0.165 |
| 17 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
-0.194 |
0.060 |
| 18 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.187 |
0.008 |
| 19 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.180 |
0.110 |
| 20 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
-0.160 |
0.051 |
| 21 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
-0.151 |
0.073 |