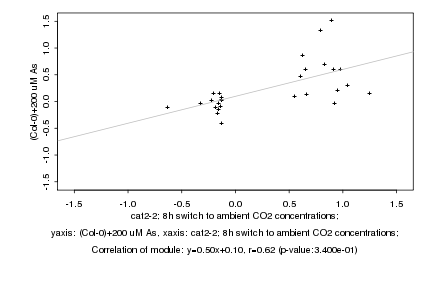
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
Log2 signal ratio
(Col-0)+200 uM As |
| 1 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
1.244 |
0.151 |
| 2 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
1.035 |
0.312 |
| 3 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.972 |
0.613 |
| 4 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.949 |
0.218 |
| 5 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.917 |
-0.021 |
| 6 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.912 |
0.614 |
| 7 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.887 |
1.533 |
| 8 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.826 |
0.703 |
| 9 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.783 |
1.331 |
| 10 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.661 |
0.146 |
| 11 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.646 |
0.616 |
| 12 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.624 |
0.868 |
| 13 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.600 |
0.473 |
| 14 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.546 |
0.111 |
| 15 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.640 |
-0.105 |
| 16 |
251292_at |
AT3G61920
|
unknown |
-0.330 |
-0.034 |
| 17 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.228 |
0.036 |
| 18 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.212 |
0.155 |
| 19 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.191 |
-0.106 |
| 20 |
244965_at |
ATCG00590
|
ORF31 |
-0.171 |
-0.221 |
| 21 |
246769_at |
AT5G27440
|
unknown |
-0.160 |
-0.027 |
| 22 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.159 |
-0.145 |
| 23 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.155 |
0.152 |
| 24 |
253319_at |
AT4G33990
|
EMB2758, embryo defective 2758 |
-0.145 |
-0.076 |
| 25 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.139 |
-0.394 |
| 26 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
-0.137 |
0.032 |
| 27 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.137 |
0.089 |
| 28 |
267527_at |
AT2G45610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.132 |
0.051 |