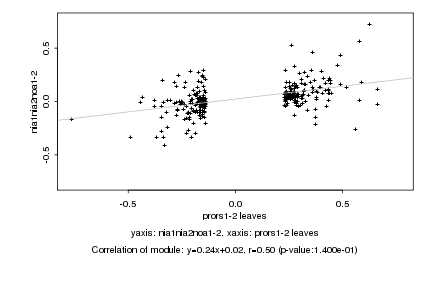
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
prors1-2 leaves |
Log2 signal ratio
nia1nia2noa1-2 |
| 1 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.659 |
0.114 |
| 2 |
247882_at |
AT5G57785
|
unknown |
0.659 |
-0.019 |
| 3 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.623 |
0.726 |
| 4 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.584 |
0.187 |
| 5 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.578 |
0.017 |
| 6 |
263515_at |
AT2G21640
|
unknown |
0.574 |
0.569 |
| 7 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.556 |
-0.258 |
| 8 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.514 |
0.135 |
| 9 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.487 |
0.167 |
| 10 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.486 |
0.435 |
| 11 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.472 |
0.338 |
| 12 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.448 |
0.081 |
| 13 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.441 |
0.198 |
| 14 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.439 |
0.174 |
| 15 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.435 |
0.078 |
| 16 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.435 |
0.222 |
| 17 |
258741_at |
AT3G05790
|
LON4, lon protease 4 |
0.432 |
0.018 |
| 18 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.432 |
0.115 |
| 19 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.431 |
0.079 |
| 20 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.431 |
0.105 |
| 21 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.430 |
0.204 |
| 22 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
0.421 |
-0.038 |
| 23 |
247410_at |
AT5G62990
|
emb1692, embryo defective 1692 |
0.418 |
0.173 |
| 24 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.416 |
0.106 |
| 25 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
0.407 |
0.220 |
| 26 |
248605_at |
AT5G49410
|
unknown |
0.405 |
0.081 |
| 27 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.398 |
0.286 |
| 28 |
247959_at |
AT5G57080
|
unknown |
0.394 |
0.138 |
| 29 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.388 |
0.134 |
| 30 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
0.380 |
0.091 |
| 31 |
258174_at |
AT3G21470
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.376 |
0.027 |
| 32 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.376 |
0.103 |
| 33 |
249965_at |
AT5G19020
|
MEF18, mitochondrial editing factor 18 |
0.374 |
0.043 |
| 34 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
0.373 |
-0.213 |
| 35 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
0.371 |
-0.143 |
| 36 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.369 |
-0.072 |
| 37 |
247616_at |
AT5G60260
|
unknown |
0.366 |
0.206 |
| 38 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.364 |
0.116 |
| 39 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.358 |
0.460 |
| 40 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.356 |
0.136 |
| 41 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.355 |
0.085 |
| 42 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.354 |
0.299 |
| 43 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.348 |
0.181 |
| 44 |
264258_at |
AT1G09220
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.340 |
0.058 |
| 45 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.336 |
0.240 |
| 46 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.332 |
-0.084 |
| 47 |
259003_at |
AT3G02010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.326 |
0.008 |
| 48 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.324 |
0.161 |
| 49 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.320 |
0.163 |
| 50 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.319 |
0.273 |
| 51 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.319 |
0.106 |
| 52 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.319 |
0.110 |
| 53 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.316 |
0.072 |
| 54 |
261124_at |
AT1G04900
|
unknown |
0.315 |
0.094 |
| 55 |
260620_at |
AT1G08070
|
EMB3102, EMBRYO DEFECTIVE 3102, OTP82, ORGANELLE TRANSCRIPT PROCESSING 82 |
0.312 |
0.032 |
| 56 |
255446_at |
AT4G02750
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.311 |
-0.015 |
| 57 |
262491_at |
AT1G21650
|
SECA2 |
0.310 |
0.060 |
| 58 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.308 |
0.176 |
| 59 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.308 |
0.212 |
| 60 |
255296_at |
AT4G04790
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.307 |
0.029 |
| 61 |
253170_at |
AT4G35130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.305 |
0.052 |
| 62 |
256775_at |
AT3G13770
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.303 |
-0.032 |
| 63 |
257057_at |
AT3G15310
|
unknown |
0.298 |
0.269 |
| 64 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
0.293 |
0.066 |
| 65 |
259285_at |
AT3G11460
|
MEF10, mitochondrial RNA editing factor 10 |
0.293 |
0.049 |
| 66 |
267110_at |
AT2G14800
|
unknown |
0.291 |
-0.045 |
| 67 |
265139_at |
AT1G51310
|
[transferases] |
0.288 |
0.047 |
| 68 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.287 |
0.068 |
| 69 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.286 |
0.066 |
| 70 |
267445_at |
AT2G33680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.285 |
0.003 |
| 71 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.285 |
0.138 |
| 72 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.284 |
0.057 |
| 73 |
259218_at |
AT3G03580
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.282 |
-0.028 |
| 74 |
258220_at |
AT3G17830
|
DJA4, DNA J protein A4 |
0.282 |
0.054 |
| 75 |
257395_at |
AT2G15630
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.282 |
0.154 |
| 76 |
263899_at |
AT2G21710
|
EMB2219, embryo defective 2219 |
0.281 |
-0.012 |
| 77 |
258157_at |
AT3G18100
|
AtMYB4R1, myb domain protein 4R1, MYB4R1, myb domain protein 4r1 |
0.280 |
0.010 |
| 78 |
254614_at |
AT4G19191
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.277 |
0.045 |
| 79 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.276 |
0.015 |
| 80 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.276 |
0.143 |
| 81 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
0.275 |
0.175 |
| 82 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.274 |
0.100 |
| 83 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.274 |
0.335 |
| 84 |
254457_at |
AT4G21170
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.273 |
-0.002 |
| 85 |
263237_at |
AT1G10610
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.273 |
-0.128 |
| 86 |
248871_at |
AT5G46680
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.272 |
0.084 |
| 87 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.272 |
0.126 |
| 88 |
250166_at |
AT5G15300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.271 |
0.013 |
| 89 |
248366_at |
AT5G52510
|
SCL8, SCARECROW-like 8 |
0.271 |
0.060 |
| 90 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.270 |
0.088 |
| 91 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.268 |
0.064 |
| 92 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.265 |
0.041 |
| 93 |
246808_at |
AT5G27110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.264 |
0.070 |
| 94 |
264946_at |
AT1G77010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.263 |
0.036 |
| 95 |
249821_at |
AT5G23690
|
[Polynucleotide adenylyltransferase family protein] |
0.262 |
0.152 |
| 96 |
267011_at |
AT2G39230
|
LOJ, LATERAL ORGAN JUNCTION |
0.261 |
0.062 |
| 97 |
247621_at |
AT5G60335
|
[Thioesterase superfamily protein] |
0.260 |
0.114 |
| 98 |
256027_at |
AT1G34160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.260 |
0.055 |
| 99 |
260522_x_at |
AT2G41730
|
unknown |
0.258 |
0.529 |
| 100 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.258 |
0.054 |
| 101 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
0.256 |
0.024 |
| 102 |
247556_at |
AT5G61040
|
unknown |
0.256 |
0.038 |
| 103 |
261715_at |
AT1G18485
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.255 |
0.048 |
| 104 |
266341_at |
AT2G01510
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.255 |
0.043 |
| 105 |
250725_at |
AT5G06400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.255 |
0.095 |
| 106 |
257979_at |
AT3G20730
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.255 |
0.020 |
| 107 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.254 |
0.072 |
| 108 |
265931_at |
AT2G18520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.253 |
0.129 |
| 109 |
248239_at |
AT5G53920
|
[ribosomal protein L11 methyltransferase-related] |
0.252 |
0.143 |
| 110 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.252 |
0.041 |
| 111 |
254208_at |
AT4G24175
|
unknown |
0.250 |
0.059 |
| 112 |
245670_at |
AT1G28210
|
ATJ1 |
0.249 |
0.057 |
| 113 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.249 |
0.052 |
| 114 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.248 |
0.182 |
| 115 |
246052_at |
AT5G08305
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.247 |
0.031 |
| 116 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.242 |
0.064 |
| 117 |
263489_at |
AT2G31830
|
5PTase14, inositol-polyphosphate 5-phosphatase 14 |
0.241 |
-0.014 |
| 118 |
266280_at |
AT2G29260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.240 |
0.078 |
| 119 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.240 |
0.075 |
| 120 |
264754_at |
AT1G61400
|
[S-locus lectin protein kinase family protein] |
0.240 |
0.031 |
| 121 |
250283_at |
AT5G13270
|
RARE1, REQUIRED FOR ACCD RNA EDITING 1 |
0.239 |
0.061 |
| 122 |
253782_at |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
0.239 |
0.041 |
| 123 |
264251_at |
AT1G09190
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.237 |
0.049 |
| 124 |
256906_at |
AT3G24000
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.235 |
0.033 |
| 125 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.235 |
0.181 |
| 126 |
252043_at |
AT3G52390
|
[TatD related DNase] |
0.235 |
0.094 |
| 127 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.234 |
-0.028 |
| 128 |
255317_at |
AT4G04180
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.234 |
0.027 |
| 129 |
262649_at |
AT1G14040
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.233 |
0.025 |
| 130 |
256837_at |
AT3G22900
|
NRPD7 |
0.232 |
0.001 |
| 131 |
264637_at |
AT1G65560
|
[Zinc-binding dehydrogenase family protein] |
0.232 |
0.077 |
| 132 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.232 |
0.015 |
| 133 |
253319_at |
AT4G33990
|
EMB2758, embryo defective 2758 |
0.231 |
0.091 |
| 134 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.231 |
0.299 |
| 135 |
252990_at |
AT4G38440
|
IYO, MINIYO |
0.230 |
0.139 |
| 136 |
255288_at |
AT4G04670
|
[Met-10+ like family protein / kelch repeat-containing protein] |
0.230 |
-0.045 |
| 137 |
255267_at |
AT4G05210
|
AtLpxD1, LpxD1, lipid X D1 |
0.228 |
0.069 |
| 138 |
251711_at |
AT3G56960
|
PIP5K4, phosphatidyl inositol monophosphate 5 kinase 4 |
0.227 |
0.043 |
| 139 |
247548_at |
AT5G61400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.227 |
-0.028 |
| 140 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.768 |
-0.160 |
| 141 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.490 |
-0.336 |
| 142 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.445 |
-0.003 |
| 143 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.437 |
0.039 |
| 144 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-0.382 |
0.012 |
| 145 |
248623_at |
AT5G49170
|
unknown |
-0.380 |
-0.042 |
| 146 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.369 |
-0.332 |
| 147 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.347 |
-0.142 |
| 148 |
256569_at |
AT3G19550
|
unknown |
-0.347 |
-0.278 |
| 149 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.345 |
-0.044 |
| 150 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.341 |
0.206 |
| 151 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.339 |
-0.329 |
| 152 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.338 |
-0.007 |
| 153 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.332 |
-0.408 |
| 154 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.325 |
-0.095 |
| 155 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.321 |
-0.235 |
| 156 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.319 |
0.018 |
| 157 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.303 |
0.016 |
| 158 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.287 |
-0.013 |
| 159 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
-0.286 |
0.181 |
| 160 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
-0.279 |
-0.006 |
| 161 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.276 |
0.143 |
| 162 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
-0.276 |
-0.129 |
| 163 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.272 |
-0.077 |
| 164 |
264998_at |
AT1G67330
|
unknown |
-0.272 |
-0.073 |
| 165 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
-0.268 |
0.249 |
| 166 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.261 |
0.003 |
| 167 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.257 |
-0.021 |
| 168 |
261895_at |
AT1G80830
|
ATNRAMP1, NRAMP1, natural resistance-associated macrophage protein 1, PMIT1 |
-0.254 |
-0.004 |
| 169 |
253238_at |
AT4G34480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.252 |
0.001 |
| 170 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.252 |
-0.012 |
| 171 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.247 |
-0.008 |
| 172 |
254351_at |
AT4G22300
|
SOBER1, SUPPRESSOR OF AVRBST-ELICITED RESISTANCE 1 |
-0.246 |
-0.073 |
| 173 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.241 |
-0.167 |
| 174 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.239 |
0.133 |
| 175 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.239 |
-0.021 |
| 176 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.236 |
0.187 |
| 177 |
252427_at |
AT3G47640
|
PYE, POPEYE |
-0.232 |
-0.155 |
| 178 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.232 |
-0.297 |
| 179 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.229 |
-0.040 |
| 180 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.226 |
-0.107 |
| 181 |
261626_at |
AT1G01990
|
unknown |
-0.223 |
-0.109 |
| 182 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.223 |
-0.270 |
| 183 |
247188_at |
AT5G65430
|
14-3-3KAPPA, 14-3-3 PROTEIN G-BOX FACTOR14 KAPPA, GF14 KAPPA, GRF8, general regulatory factor 8 |
-0.220 |
-0.099 |
| 184 |
251638_at |
AT3G57490
|
[Ribosomal protein S5 family protein] |
-0.218 |
0.061 |
| 185 |
246911_at |
AT5G25810
|
tny, TINY |
-0.217 |
-0.146 |
| 186 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.216 |
-0.160 |
| 187 |
260841_at |
AT1G29195
|
unknown |
-0.214 |
0.285 |
| 188 |
247522_at |
AT5G61340
|
unknown |
-0.212 |
-0.108 |
| 189 |
249204_at |
AT5G42570
|
[B-cell receptor-associated 31-like] |
-0.211 |
-0.011 |
| 190 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
-0.210 |
0.006 |
| 191 |
267019_at |
AT2G39130
|
[Transmembrane amino acid transporter family protein] |
-0.207 |
-0.059 |
| 192 |
258078_at |
AT3G25870
|
unknown |
-0.207 |
0.027 |
| 193 |
254057_at |
AT4G25170
|
[Uncharacterised conserved protein (UCP012943)] |
-0.205 |
-0.336 |
| 194 |
262313_at |
AT1G70900
|
unknown |
-0.201 |
-0.018 |
| 195 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.201 |
0.023 |
| 196 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.200 |
0.107 |
| 197 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
-0.199 |
-0.197 |
| 198 |
251616_at |
AT3G57990
|
unknown |
-0.197 |
-0.087 |
| 199 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.197 |
-0.082 |
| 200 |
250278_at |
AT5G12860
|
DiT1, dicarboxylate transporter 1 |
-0.195 |
0.066 |
| 201 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.190 |
-0.033 |
| 202 |
253198_at |
AT4G35360
|
[Uncharacterised conserved protein (UCP030210)] |
-0.190 |
-0.099 |
| 203 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.190 |
0.059 |
| 204 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.187 |
-0.296 |
| 205 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.186 |
-0.022 |
| 206 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.185 |
0.077 |
| 207 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.184 |
-0.081 |
| 208 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.183 |
0.062 |
| 209 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.181 |
0.030 |
| 210 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.181 |
0.137 |
| 211 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.179 |
-0.002 |
| 212 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.178 |
-0.099 |
| 213 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.177 |
0.081 |
| 214 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
-0.176 |
0.189 |
| 215 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
-0.175 |
0.279 |
| 216 |
248951_at |
AT5G45550
|
MOB1-like, MOB1-like |
-0.173 |
0.003 |
| 217 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.172 |
0.044 |
| 218 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.171 |
0.025 |
| 219 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.171 |
-0.128 |
| 220 |
251574_at |
AT3G58100
|
PDCB5, plasmodesmata callose-binding protein 5 |
-0.170 |
-0.019 |
| 221 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.170 |
-0.102 |
| 222 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
-0.169 |
0.099 |
| 223 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.169 |
-0.084 |
| 224 |
247762_at |
AT5G59170
|
[Proline-rich extensin-like family protein] |
-0.167 |
0.018 |
| 225 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.167 |
-0.083 |
| 226 |
258213_at |
AT3G17950
|
unknown |
-0.167 |
-0.046 |
| 227 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.166 |
0.042 |
| 228 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.165 |
-0.099 |
| 229 |
251012_at |
AT5G02580
|
unknown |
-0.165 |
-0.026 |
| 230 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.164 |
-0.159 |
| 231 |
247715_at |
AT5G59360
|
unknown |
-0.164 |
-0.052 |
| 232 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.160 |
0.185 |
| 233 |
262434_at |
AT1G47670
|
[Transmembrane amino acid transporter family protein] |
-0.160 |
-0.011 |
| 234 |
261292_at |
AT1G36940
|
unknown |
-0.158 |
-0.001 |
| 235 |
261415_at |
AT1G07750
|
[RmlC-like cupins superfamily protein] |
-0.158 |
0.027 |
| 236 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
-0.157 |
0.252 |
| 237 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.156 |
0.031 |
| 238 |
252307_at |
AT3G49280
|
unknown |
-0.156 |
-0.034 |
| 239 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.156 |
-0.118 |
| 240 |
266609_at |
AT2G35510
|
SRO1, similar to RCD one 1 |
-0.155 |
-0.097 |
| 241 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.154 |
-0.045 |
| 242 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.154 |
-0.133 |
| 243 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.154 |
0.240 |
| 244 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.154 |
-0.014 |
| 245 |
248511_at |
AT5G50375
|
CPI1, cyclopropyl isomerase |
-0.154 |
-0.081 |
| 246 |
248121_at |
AT5G54690
|
GAUT12, galacturonosyltransferase 12, IRX8, IRREGULAR XYLEM 8, LGT6 |
-0.153 |
-0.016 |
| 247 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
-0.152 |
0.014 |
| 248 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.152 |
-0.038 |
| 249 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.152 |
0.005 |
| 250 |
258911_at |
AT3G06470
|
[GNS1/SUR4 membrane protein family] |
-0.151 |
0.031 |
| 251 |
255300_at |
AT4G04870
|
CLS, cardiolipin synthase |
-0.151 |
0.063 |
| 252 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.151 |
0.293 |
| 253 |
253437_at |
AT4G32460
|
unknown |
-0.151 |
0.041 |
| 254 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.151 |
0.233 |
| 255 |
267055_at |
AT2G38360
|
PRA1.B4, prenylated RAB acceptor 1.B4 |
-0.150 |
-0.053 |
| 256 |
248144_at |
AT5G54800
|
ATGPT1, ARABIDOPSIS GLUCOSE 6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 1, GPT1, glucose 6-phosphate/phosphate translocator 1 |
-0.150 |
0.149 |
| 257 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.149 |
-0.040 |
| 258 |
265791_at |
AT2G01280
|
MEE65, maternal effect embryo arrest 65 |
-0.148 |
-0.006 |
| 259 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.148 |
-0.034 |
| 260 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.148 |
-0.146 |
| 261 |
253343_at |
AT4G33540
|
[metallo-beta-lactamase family protein] |
-0.148 |
0.112 |
| 262 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.148 |
0.028 |
| 263 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.147 |
-0.097 |
| 264 |
263197_at |
AT1G53930
|
[Ubiquitin-like superfamily protein] |
-0.147 |
-0.046 |
| 265 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.146 |
-0.057 |
| 266 |
251236_at |
AT3G62380
|
unknown |
-0.146 |
-0.018 |
| 267 |
249951_at |
AT5G18930
|
BUD2, BUSHY AND DWARF 2, SAMDC4 |
-0.145 |
0.023 |
| 268 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
-0.145 |
-0.087 |
| 269 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.145 |
0.014 |
| 270 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.144 |
-0.201 |
| 271 |
252758_at |
AT3G42710
|
[pseudogene] |
-0.144 |
-0.021 |
| 272 |
265944_at |
AT2G19640
|
ASHR2, ASH1-related 2, SDG39, SET DOMAIN PROTEIN 39 |
-0.144 |
0.091 |
| 273 |
251796_at |
AT3G55360
|
ATTSC13, CER10, ECERIFERUM 10, ECR, ENOYL-COA REDUCTASE, TSC13 |
-0.144 |
0.032 |
| 274 |
246757_at |
AT5G27940
|
WPP3, WPP domain protein 3 |
-0.143 |
-0.038 |
| 275 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.143 |
0.208 |
| 276 |
257213_at |
AT3G15020
|
mMDH2, mitochondrial malate dehydrogenase 2 |
-0.142 |
0.006 |
| 277 |
253110_at |
AT4G35930
|
AtFBS4, FBS4, F-box stress induced 4 |
-0.141 |
-0.017 |