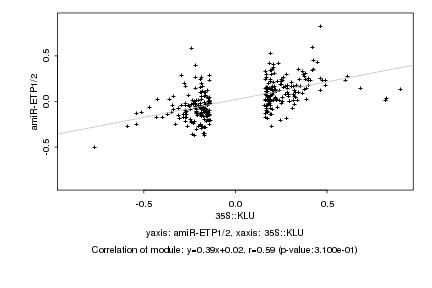
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::KLU |
Log2 signal ratio
amiR-ETP1/2 |
| 1 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.899 |
0.136 |
| 2 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
0.821 |
0.043 |
| 3 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
0.819 |
0.017 |
| 4 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.678 |
0.151 |
| 5 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.611 |
0.285 |
| 6 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.597 |
0.235 |
| 7 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.490 |
0.240 |
| 8 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.488 |
0.185 |
| 9 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.474 |
0.237 |
| 10 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.460 |
0.128 |
| 11 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.459 |
0.826 |
| 12 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.459 |
0.253 |
| 13 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.446 |
0.432 |
| 14 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.425 |
0.355 |
| 15 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.424 |
0.457 |
| 16 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.419 |
0.347 |
| 17 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.419 |
0.598 |
| 18 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.405 |
0.233 |
| 19 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.395 |
0.235 |
| 20 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.395 |
0.178 |
| 21 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.394 |
0.258 |
| 22 |
263158_at |
AT1G54160
|
NF-YA5, nuclear factor Y, subunit A5, NFYA5, NUCLEAR FACTOR Y A5 |
0.385 |
0.150 |
| 23 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.384 |
0.028 |
| 24 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.380 |
0.246 |
| 25 |
252888_at |
AT4G39210
|
APL3 |
0.379 |
0.315 |
| 26 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.372 |
0.278 |
| 27 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.372 |
0.137 |
| 28 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.369 |
0.301 |
| 29 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.366 |
0.092 |
| 30 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.364 |
0.330 |
| 31 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.351 |
0.168 |
| 32 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.347 |
0.244 |
| 33 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.341 |
0.352 |
| 34 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.340 |
0.105 |
| 35 |
262452_at |
AT1G11210
|
unknown |
0.338 |
0.022 |
| 36 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.329 |
0.177 |
| 37 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.326 |
0.045 |
| 38 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.325 |
0.102 |
| 39 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.322 |
0.124 |
| 40 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.319 |
-0.029 |
| 41 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.318 |
0.057 |
| 42 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.316 |
0.082 |
| 43 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.314 |
0.019 |
| 44 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.312 |
0.174 |
| 45 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
0.310 |
-0.074 |
| 46 |
256924_at |
AT3G29590
|
AT5MAT |
0.305 |
0.210 |
| 47 |
256789_at |
AT3G13672
|
SINA2 |
0.301 |
0.170 |
| 48 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.294 |
0.001 |
| 49 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.290 |
0.004 |
| 50 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.289 |
0.084 |
| 51 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.287 |
0.058 |
| 52 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.284 |
0.105 |
| 53 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.283 |
0.183 |
| 54 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.279 |
0.150 |
| 55 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
0.278 |
0.299 |
| 56 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.276 |
-0.177 |
| 57 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.269 |
0.082 |
| 58 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.268 |
0.169 |
| 59 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.262 |
0.162 |
| 60 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.257 |
0.272 |
| 61 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.255 |
0.046 |
| 62 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.253 |
0.006 |
| 63 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.252 |
0.231 |
| 64 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.250 |
0.252 |
| 65 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.249 |
-0.011 |
| 66 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.248 |
0.230 |
| 67 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.246 |
0.197 |
| 68 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.245 |
0.192 |
| 69 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.241 |
-0.200 |
| 70 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.237 |
0.019 |
| 71 |
256114_at |
AT1G16850
|
unknown |
0.237 |
0.128 |
| 72 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.235 |
0.418 |
| 73 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.229 |
-0.065 |
| 74 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
0.225 |
0.229 |
| 75 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.222 |
0.120 |
| 76 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.221 |
0.024 |
| 77 |
265539_at |
AT2G15830
|
unknown |
0.220 |
0.016 |
| 78 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.219 |
0.022 |
| 79 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.218 |
0.138 |
| 80 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.215 |
0.048 |
| 81 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.214 |
0.043 |
| 82 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
0.214 |
0.035 |
| 83 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
0.212 |
0.314 |
| 84 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.210 |
0.170 |
| 85 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.210 |
0.137 |
| 86 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.207 |
0.100 |
| 87 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.207 |
0.381 |
| 88 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.206 |
0.411 |
| 89 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.205 |
0.218 |
| 90 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.204 |
0.017 |
| 91 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.204 |
0.099 |
| 92 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.201 |
-0.086 |
| 93 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.201 |
0.175 |
| 94 |
265276_at |
AT2G28400
|
unknown |
0.199 |
0.359 |
| 95 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.199 |
0.164 |
| 96 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.199 |
0.127 |
| 97 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.197 |
0.123 |
| 98 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.197 |
0.126 |
| 99 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
0.197 |
0.086 |
| 100 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.196 |
0.344 |
| 101 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
0.196 |
0.138 |
| 102 |
258198_at |
AT3G14020
|
NF-YA6, nuclear factor Y, subunit A6 |
0.194 |
-0.133 |
| 103 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
0.194 |
0.301 |
| 104 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.192 |
0.071 |
| 105 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.192 |
-0.268 |
| 106 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
0.191 |
-0.066 |
| 107 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
0.191 |
0.068 |
| 108 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.190 |
-0.134 |
| 109 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
0.189 |
0.026 |
| 110 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.189 |
0.004 |
| 111 |
257053_at |
AT3G15210
|
ATERF-4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ATERF4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ERF4, ethylene responsive element binding factor 4, RAP2.5, RELATED TO AP2 5 |
0.189 |
0.350 |
| 112 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
0.188 |
0.065 |
| 113 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.187 |
0.023 |
| 114 |
254178_at |
AT4G23880
|
unknown |
0.187 |
0.533 |
| 115 |
260345_at |
AT1G69270
|
AtRPK1, RPK1, receptor-like protein kinase 1 |
0.187 |
0.252 |
| 116 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.186 |
0.076 |
| 117 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.186 |
0.220 |
| 118 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
0.186 |
-0.038 |
| 119 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.185 |
-0.016 |
| 120 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
0.184 |
0.020 |
| 121 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.184 |
0.417 |
| 122 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
0.182 |
0.019 |
| 123 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
0.181 |
0.143 |
| 124 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.180 |
0.203 |
| 125 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.178 |
0.051 |
| 126 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.177 |
-0.020 |
| 127 |
248969_at |
AT5G45310
|
unknown |
0.176 |
0.008 |
| 128 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.174 |
0.110 |
| 129 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.174 |
0.057 |
| 130 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.173 |
-0.183 |
| 131 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.172 |
0.032 |
| 132 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.172 |
-0.118 |
| 133 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.172 |
-0.034 |
| 134 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.171 |
0.159 |
| 135 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.171 |
0.136 |
| 136 |
249303_at |
AT5G41460
|
unknown |
0.170 |
-0.104 |
| 137 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
0.170 |
0.141 |
| 138 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.169 |
0.069 |
| 139 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.169 |
-0.053 |
| 140 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.169 |
0.115 |
| 141 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.168 |
-0.037 |
| 142 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.168 |
-0.070 |
| 143 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.168 |
0.117 |
| 144 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.167 |
-0.098 |
| 145 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.167 |
0.160 |
| 146 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.166 |
0.097 |
| 147 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
0.165 |
0.306 |
| 148 |
258935_at |
AT3G10120
|
unknown |
0.165 |
0.271 |
| 149 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.164 |
-0.123 |
| 150 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
0.163 |
0.028 |
| 151 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.163 |
0.340 |
| 152 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.161 |
0.083 |
| 153 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.161 |
0.013 |
| 154 |
266396_at |
AT2G38790
|
unknown |
0.160 |
0.252 |
| 155 |
260262_at |
AT1G68470
|
[Exostosin family protein] |
0.160 |
-0.168 |
| 156 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.159 |
0.147 |
| 157 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
0.158 |
-0.036 |
| 158 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.771 |
-0.496 |
| 159 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.591 |
-0.267 |
| 160 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.542 |
-0.242 |
| 161 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.541 |
-0.123 |
| 162 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.515 |
-0.114 |
| 163 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.471 |
-0.058 |
| 164 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.433 |
-0.166 |
| 165 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.427 |
0.027 |
| 166 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.401 |
-0.168 |
| 167 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.372 |
-0.139 |
| 168 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.361 |
0.025 |
| 169 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.353 |
-0.119 |
| 170 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.346 |
-0.039 |
| 171 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.342 |
-0.077 |
| 172 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.339 |
0.060 |
| 173 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.330 |
-0.243 |
| 174 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.316 |
-0.147 |
| 175 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.316 |
-0.071 |
| 176 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.312 |
-0.072 |
| 177 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.307 |
-0.178 |
| 178 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.299 |
0.296 |
| 179 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.295 |
-0.037 |
| 180 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.287 |
-0.136 |
| 181 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.285 |
-0.167 |
| 182 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.279 |
0.206 |
| 183 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.278 |
0.166 |
| 184 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.277 |
-0.091 |
| 185 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.276 |
-0.019 |
| 186 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.276 |
-0.115 |
| 187 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.274 |
-0.209 |
| 188 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.274 |
-0.174 |
| 189 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.274 |
-0.033 |
| 190 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.272 |
-0.142 |
| 191 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.271 |
-0.104 |
| 192 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.268 |
-0.173 |
| 193 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.263 |
-0.273 |
| 194 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.258 |
-0.033 |
| 195 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.257 |
0.075 |
| 196 |
258468_at |
AT3G06070
|
unknown |
-0.251 |
-0.048 |
| 197 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.250 |
-0.079 |
| 198 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.250 |
-0.190 |
| 199 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.243 |
0.582 |
| 200 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.242 |
-0.027 |
| 201 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.242 |
-0.217 |
| 202 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.240 |
-0.224 |
| 203 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.236 |
0.016 |
| 204 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.235 |
-0.359 |
| 205 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.233 |
-0.011 |
| 206 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.233 |
-0.029 |
| 207 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.233 |
-0.235 |
| 208 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.228 |
-0.371 |
| 209 |
261772_at |
AT1G76240
|
unknown |
-0.226 |
-0.211 |
| 210 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.225 |
0.004 |
| 211 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.222 |
0.265 |
| 212 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
-0.221 |
-0.051 |
| 213 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.220 |
-0.016 |
| 214 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
-0.220 |
0.148 |
| 215 |
257551_at |
AT3G13270
|
unknown |
-0.220 |
0.404 |
| 216 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.220 |
-0.026 |
| 217 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.220 |
-0.071 |
| 218 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.218 |
-0.113 |
| 219 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.218 |
0.020 |
| 220 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.215 |
-0.081 |
| 221 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.215 |
-0.122 |
| 222 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.215 |
-0.106 |
| 223 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.214 |
-0.298 |
| 224 |
260012_at |
AT1G67865
|
unknown |
-0.214 |
-0.106 |
| 225 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.210 |
-0.129 |
| 226 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.208 |
-0.153 |
| 227 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.207 |
-0.267 |
| 228 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.206 |
-0.037 |
| 229 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.206 |
0.014 |
| 230 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.200 |
-0.057 |
| 231 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.196 |
0.104 |
| 232 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.195 |
-0.141 |
| 233 |
256674_at |
AT3G52360
|
unknown |
-0.195 |
-0.050 |
| 234 |
247754_at |
AT5G59080
|
unknown |
-0.195 |
-0.243 |
| 235 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.195 |
0.162 |
| 236 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.194 |
-0.043 |
| 237 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.194 |
0.242 |
| 238 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.193 |
-0.014 |
| 239 |
249932_at |
AT5G22390
|
unknown |
-0.192 |
-0.255 |
| 240 |
258275_at |
AT3G15760
|
unknown |
-0.191 |
-0.042 |
| 241 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
-0.190 |
-0.162 |
| 242 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.190 |
-0.115 |
| 243 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
-0.189 |
0.026 |
| 244 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.189 |
-0.293 |
| 245 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.189 |
-0.127 |
| 246 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.189 |
0.059 |
| 247 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.188 |
-0.067 |
| 248 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.188 |
-0.077 |
| 249 |
246200_at |
AT4G37240
|
unknown |
-0.188 |
0.208 |
| 250 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.187 |
0.103 |
| 251 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.186 |
0.265 |
| 252 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.185 |
-0.269 |
| 253 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
-0.185 |
0.120 |
| 254 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.183 |
0.169 |
| 255 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.181 |
-0.267 |
| 256 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.180 |
0.171 |
| 257 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.180 |
-0.147 |
| 258 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.179 |
-0.342 |
| 259 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.178 |
-0.056 |
| 260 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.177 |
-0.164 |
| 261 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
-0.175 |
-0.095 |
| 262 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.174 |
-0.070 |
| 263 |
261247_at |
AT1G20070
|
unknown |
-0.173 |
-0.350 |
| 264 |
260004_at |
AT1G67860
|
unknown |
-0.173 |
-0.009 |
| 265 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.173 |
0.057 |
| 266 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.173 |
-0.367 |
| 267 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.170 |
-0.277 |
| 268 |
262801_at |
AT1G21010
|
unknown |
-0.170 |
0.100 |
| 269 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.169 |
-0.119 |
| 270 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.169 |
-0.191 |
| 271 |
256578_at |
AT3G28200
|
[Peroxidase superfamily protein] |
-0.167 |
0.067 |
| 272 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.167 |
0.107 |
| 273 |
264521_at |
AT1G10020
|
unknown |
-0.167 |
-0.073 |
| 274 |
266968_at |
AT2G39360
|
[Protein kinase superfamily protein] |
-0.165 |
-0.027 |
| 275 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.165 |
-0.089 |
| 276 |
249378_at |
AT5G40450
|
unknown |
-0.164 |
-0.283 |
| 277 |
263632_at |
AT2G04795
|
unknown |
-0.164 |
-0.083 |
| 278 |
266824_at |
AT2G22800
|
HAT9 |
-0.162 |
-0.117 |
| 279 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.162 |
-0.079 |
| 280 |
256025_at |
AT1G58370
|
ATXYN1, ARABIDOPSIS THALIANA XYLANASE 1, RXF12 |
-0.161 |
-0.155 |
| 281 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.161 |
-0.103 |
| 282 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.159 |
-0.216 |
| 283 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.158 |
-0.041 |
| 284 |
245353_at |
AT4G16000
|
unknown |
-0.157 |
-0.122 |
| 285 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.156 |
-0.207 |
| 286 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.156 |
-0.157 |
| 287 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.155 |
-0.135 |
| 288 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.154 |
-0.138 |
| 289 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.154 |
-0.146 |
| 290 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.153 |
0.129 |
| 291 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.153 |
0.050 |
| 292 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.151 |
-0.027 |
| 293 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.151 |
-0.017 |
| 294 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.150 |
-0.118 |
| 295 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.149 |
-0.203 |
| 296 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.149 |
-0.028 |
| 297 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.148 |
0.004 |
| 298 |
263650_at |
AT1G04360
|
[RING/U-box superfamily protein] |
-0.148 |
-0.031 |
| 299 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.148 |
0.040 |
| 300 |
267459_at |
AT2G33850
|
unknown |
-0.147 |
-0.006 |
| 301 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.146 |
-0.098 |
| 302 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.146 |
0.008 |
| 303 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.145 |
0.233 |
| 304 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.145 |
0.005 |
| 305 |
261193_at |
AT1G32920
|
unknown |
-0.144 |
0.293 |
| 306 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.144 |
-0.146 |
| 307 |
257502_at |
AT1G78110
|
unknown |
-0.144 |
0.050 |
| 308 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
-0.144 |
0.017 |
| 309 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.144 |
-0.246 |
| 310 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.144 |
-0.204 |
| 311 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-0.143 |
-0.136 |
| 312 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
-0.141 |
-0.018 |
| 313 |
250223_at |
AT5G14070
|
ROXY2 |
-0.141 |
-0.205 |