|
probeID |
AGICode |
Annotation |
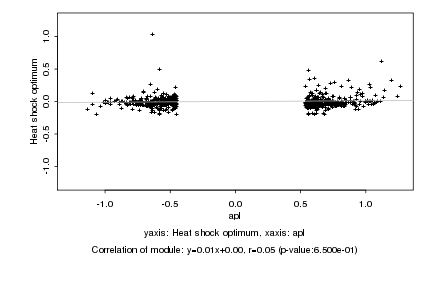
Log2 signal ratio
apl |
Log2 signal ratio
Heat shock optimum |
| 1 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
1.263 |
0.239 |
| 2 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.239 |
0.092 |
| 3 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
1.196 |
0.330 |
| 4 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
1.137 |
0.183 |
| 5 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.130 |
0.062 |
| 6 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
1.119 |
0.628 |
| 7 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.111 |
0.010 |
| 8 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
1.085 |
0.006 |
| 9 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
1.079 |
-0.013 |
| 10 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
1.070 |
0.096 |
| 11 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
1.064 |
-0.018 |
| 12 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
1.062 |
-0.012 |
| 13 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
1.060 |
-0.024 |
| 14 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
1.058 |
-0.031 |
| 15 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
1.045 |
0.026 |
| 16 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
1.036 |
-0.034 |
| 17 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.034 |
0.218 |
| 18 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
1.030 |
0.001 |
| 19 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
1.026 |
0.007 |
| 20 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
1.024 |
0.268 |
| 21 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
1.018 |
-0.033 |
| 22 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
1.012 |
0.013 |
| 23 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
1.011 |
0.031 |
| 24 |
246857_at |
AT5G25920
|
unknown |
0.996 |
-0.010 |
| 25 |
257595_at |
AT3G24750
|
unknown |
0.992 |
0.005 |
| 26 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.990 |
0.004 |
| 27 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.981 |
-0.066 |
| 28 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.973 |
0.092 |
| 29 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.967 |
0.137 |
| 30 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
0.959 |
0.005 |
| 31 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.953 |
0.111 |
| 32 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.945 |
-0.039 |
| 33 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.945 |
0.193 |
| 34 |
246781_at |
AT5G27350
|
SFP1 |
0.942 |
-0.109 |
| 35 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.929 |
0.139 |
| 36 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
0.928 |
0.040 |
| 37 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.925 |
0.094 |
| 38 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.921 |
-0.064 |
| 39 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.919 |
-0.122 |
| 40 |
245226_at |
AT3G29970
|
[B12D protein] |
0.915 |
-0.051 |
| 41 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.913 |
-0.016 |
| 42 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
0.911 |
-0.013 |
| 43 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
0.908 |
-0.017 |
| 44 |
245483_at |
AT4G16190
|
[Papain family cysteine protease] |
0.899 |
0.015 |
| 45 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.893 |
-0.045 |
| 46 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.890 |
0.225 |
| 47 |
265560_at |
AT2G05520
|
ATGRP-3, GLYCINE-RICH PROTEIN 3, ATGRP3, ARABIDOPSIS GLYCINE RICH PROTEIN 3, GRP-3, glycine-rich protein 3, GRP3, GLYCINE-RICH PROTEIN 3 |
0.882 |
0.025 |
| 48 |
255435_at |
AT4G03280
|
PETC, photosynthetic electron transfer C, PGR1, PROTON GRADIENT REGULATION 1 |
0.877 |
-0.010 |
| 49 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.876 |
0.075 |
| 50 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.875 |
0.065 |
| 51 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.867 |
-0.009 |
| 52 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.862 |
0.335 |
| 53 |
247320_at |
AT5G64040
|
PSAN |
0.851 |
-0.009 |
| 54 |
262864_at |
AT1G64920
|
[UDP-Glycosyltransferase superfamily protein] |
0.848 |
0.017 |
| 55 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
0.844 |
-0.032 |
| 56 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.842 |
0.027 |
| 57 |
256044_at |
AT1G07160
|
[Protein phosphatase 2C family protein] |
0.835 |
-0.069 |
| 58 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.831 |
-0.046 |
| 59 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
0.831 |
-0.034 |
| 60 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.830 |
0.055 |
| 61 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.828 |
-0.025 |
| 62 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.825 |
-0.067 |
| 63 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.824 |
-0.037 |
| 64 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
0.821 |
-0.014 |
| 65 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.814 |
-0.009 |
| 66 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.812 |
0.241 |
| 67 |
258178_at |
AT3G21680
|
unknown |
0.812 |
0.007 |
| 68 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.811 |
-0.044 |
| 69 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
0.809 |
-0.008 |
| 70 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.808 |
-0.024 |
| 71 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.806 |
-0.062 |
| 72 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.805 |
-0.051 |
| 73 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
0.803 |
-0.049 |
| 74 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.801 |
-0.020 |
| 75 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.798 |
0.048 |
| 76 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.796 |
0.069 |
| 77 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
0.787 |
-0.004 |
| 78 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.784 |
-0.053 |
| 79 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.783 |
-0.044 |
| 80 |
252218_at |
AT3G50150
|
unknown |
0.781 |
0.014 |
| 81 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.781 |
-0.038 |
| 82 |
261197_at |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
0.779 |
-0.022 |
| 83 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.774 |
-0.042 |
| 84 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.771 |
-0.062 |
| 85 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.766 |
-0.050 |
| 86 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.764 |
-0.070 |
| 87 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
0.764 |
-0.004 |
| 88 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.763 |
-0.056 |
| 89 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
0.762 |
-0.043 |
| 90 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.761 |
-0.015 |
| 91 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.758 |
-0.044 |
| 92 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.755 |
0.306 |
| 93 |
255653_at |
AT4G00960
|
[Protein kinase superfamily protein] |
0.754 |
-0.031 |
| 94 |
254205_at |
AT4G24170
|
[ATP binding microtubule motor family protein] |
0.751 |
0.005 |
| 95 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.749 |
0.091 |
| 96 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
0.748 |
-0.034 |
| 97 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
0.744 |
-0.018 |
| 98 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
0.744 |
-0.081 |
| 99 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.740 |
0.088 |
| 100 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
0.740 |
-0.013 |
| 101 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.738 |
0.031 |
| 102 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.738 |
-0.026 |
| 103 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.736 |
-0.054 |
| 104 |
260495_at |
AT2G41810
|
unknown |
0.733 |
-0.029 |
| 105 |
264837_at |
AT1G03600
|
PSB27 |
0.732 |
-0.009 |
| 106 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.729 |
0.286 |
| 107 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
0.726 |
-0.059 |
| 108 |
248407_at |
AT5G51500
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.726 |
-0.014 |
| 109 |
247778_at |
AT5G58750
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.725 |
0.024 |
| 110 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
0.723 |
0.001 |
| 111 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.723 |
-0.043 |
| 112 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.723 |
-0.028 |
| 113 |
258364_at |
AT3G14225
|
GLIP4, GDSL-motif lipase 4 |
0.722 |
-0.008 |
| 114 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.717 |
0.019 |
| 115 |
253823_at |
AT4G28030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.714 |
-0.025 |
| 116 |
253096_at |
AT4G37330
|
CYP81D4, cytochrome P450, family 81, subfamily D, polypeptide 4 |
0.712 |
-0.121 |
| 117 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
0.711 |
-0.030 |
| 118 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.710 |
0.061 |
| 119 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
0.710 |
0.012 |
| 120 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.708 |
-0.104 |
| 121 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.708 |
0.015 |
| 122 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.707 |
0.022 |
| 123 |
252490_at |
AT3G46720
|
[UDP-Glycosyltransferase superfamily protein] |
0.707 |
-0.043 |
| 124 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
0.706 |
0.026 |
| 125 |
256811_at |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
0.706 |
-0.036 |
| 126 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.706 |
-0.017 |
| 127 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.705 |
-0.035 |
| 128 |
266000_at |
AT2G24180
|
CYP71B6, cytochrome p450 71b6 |
0.698 |
-0.031 |
| 129 |
262638_at |
AT1G06650
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.698 |
-0.038 |
| 130 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.696 |
0.030 |
| 131 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
0.696 |
-0.022 |
| 132 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.695 |
0.138 |
| 133 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.691 |
0.000 |
| 134 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.689 |
-0.044 |
| 135 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.688 |
0.125 |
| 136 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
0.688 |
-0.040 |
| 137 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.687 |
0.033 |
| 138 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.686 |
0.210 |
| 139 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
0.685 |
-0.080 |
| 140 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.684 |
-0.125 |
| 141 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.682 |
-0.198 |
| 142 |
261572_at |
AT1G01170
|
unknown |
0.681 |
0.008 |
| 143 |
260752_at |
AT1G49030
|
[PLAC8 family protein] |
0.679 |
-0.022 |
| 144 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.672 |
0.067 |
| 145 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.670 |
-0.177 |
| 146 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.669 |
-0.032 |
| 147 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.668 |
-0.047 |
| 148 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.667 |
0.118 |
| 149 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
0.667 |
-0.027 |
| 150 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
0.666 |
0.081 |
| 151 |
258094_at |
AT3G14690
|
CYP72A15, cytochrome P450, family 72, subfamily A, polypeptide 15 |
0.665 |
-0.076 |
| 152 |
252217_at |
AT3G50140
|
unknown |
0.664 |
-0.038 |
| 153 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.664 |
-0.026 |
| 154 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.662 |
0.016 |
| 155 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.661 |
0.020 |
| 156 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.661 |
-0.077 |
| 157 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.657 |
-0.057 |
| 158 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.657 |
-0.025 |
| 159 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.657 |
-0.069 |
| 160 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
0.656 |
-0.070 |
| 161 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.655 |
0.023 |
| 162 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.652 |
0.044 |
| 163 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.651 |
0.008 |
| 164 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.649 |
0.036 |
| 165 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
0.648 |
-0.014 |
| 166 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.648 |
0.139 |
| 167 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.647 |
0.009 |
| 168 |
247813_at |
AT5G58330
|
[lactate/malate dehydrogenase family protein] |
0.646 |
-0.039 |
| 169 |
264092_at |
AT1G79040
|
PSBR, photosystem II subunit R |
0.645 |
-0.010 |
| 170 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.643 |
0.131 |
| 171 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
0.642 |
-0.018 |
| 172 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.641 |
-0.053 |
| 173 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
0.641 |
-0.021 |
| 174 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
0.639 |
-0.039 |
| 175 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.637 |
0.034 |
| 176 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
0.636 |
-0.079 |
| 177 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.633 |
0.248 |
| 178 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.632 |
0.071 |
| 179 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.632 |
-0.051 |
| 180 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.631 |
-0.023 |
| 181 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
0.631 |
0.010 |
| 182 |
247073_at |
AT5G66570
|
MSP-1, MANGANESE-STABILIZING PROTEIN 1, OE33, OXYGEN EVOLVING COMPLEX 33 KILODALTON PROTEIN, OEE1, 33 KDA OXYGEN EVOLVING POLYPEPTIDE 1, OEE33, OXYGEN EVOLVING ENHANCER PROTEIN 33, PSBO-1, PS II OXYGEN-EVOLVING COMPLEX 1, PSBO1, PS II oxygen-evolving complex 1 |
0.630 |
0.001 |
| 183 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.629 |
0.063 |
| 184 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
0.628 |
-0.002 |
| 185 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.627 |
-0.119 |
| 186 |
265438_at |
AT2G20970
|
unknown |
0.627 |
-0.014 |
| 187 |
258172_at |
AT3G21620
|
[ERD (early-responsive to dehydration stress) family protein] |
0.627 |
-0.062 |
| 188 |
245767_at |
AT1G33612
|
[Leucine-rich repeat (LRR) family protein] |
0.626 |
-0.033 |
| 189 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.625 |
0.067 |
| 190 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.625 |
-0.019 |
| 191 |
253211_at |
AT4G34880
|
[Amidase family protein] |
0.625 |
-0.048 |
| 192 |
245560_at |
AT4G15480
|
UGT84A1 |
0.624 |
-0.064 |
| 193 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.624 |
-0.043 |
| 194 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.622 |
0.029 |
| 195 |
247193_at |
AT5G65380
|
[MATE efflux family protein] |
0.620 |
-0.024 |
| 196 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.620 |
-0.172 |
| 197 |
256362_at |
AT1G66450
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.618 |
0.005 |
| 198 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.618 |
-0.133 |
| 199 |
247882_at |
AT5G57785
|
unknown |
0.617 |
0.171 |
| 200 |
246063_at |
AT5G19340
|
unknown |
0.616 |
0.043 |
| 201 |
260472_at |
AT1G10990
|
unknown |
0.615 |
0.175 |
| 202 |
265611_at |
AT2G25510
|
unknown |
0.612 |
-0.016 |
| 203 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.610 |
0.035 |
| 204 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.608 |
0.090 |
| 205 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.608 |
-0.101 |
| 206 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.608 |
-0.014 |
| 207 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
0.607 |
-0.081 |
| 208 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.606 |
-0.190 |
| 209 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.606 |
-0.028 |
| 210 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.606 |
-0.013 |
| 211 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.603 |
0.369 |
| 212 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.603 |
-0.055 |
| 213 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
0.601 |
-0.006 |
| 214 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.601 |
0.074 |
| 215 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.599 |
0.052 |
| 216 |
261275_at |
AT1G26700
|
ATMLO14, MILDEW RESISTANCE LOCUS O 14, MLO14, MILDEW RESISTANCE LOCUS O 14 |
0.598 |
-0.051 |
| 217 |
253044_at |
AT4G37290
|
unknown |
0.597 |
0.035 |
| 218 |
263709_at |
AT1G09310
|
unknown |
0.593 |
-0.009 |
| 219 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.593 |
-0.078 |
| 220 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
0.593 |
-0.009 |
| 221 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.592 |
0.026 |
| 222 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.592 |
-0.060 |
| 223 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.590 |
0.104 |
| 224 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
0.589 |
-0.035 |
| 225 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.589 |
-0.047 |
| 226 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.589 |
0.031 |
| 227 |
245061_at |
AT2G39730
|
RCA, rubisco activase |
0.588 |
-0.018 |
| 228 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
0.588 |
0.137 |
| 229 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.587 |
-0.083 |
| 230 |
250135_at |
AT5G15360
|
unknown |
0.586 |
-0.061 |
| 231 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.586 |
-0.178 |
| 232 |
265329_at |
AT2G18450
|
SDH1-2, succinate dehydrogenase 1-2 |
0.585 |
-0.051 |
| 233 |
247475_at |
AT5G62310
|
IRE, INCOMPLETE ROOT HAIR ELONGATION |
0.585 |
-0.009 |
| 234 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
0.585 |
0.004 |
| 235 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
0.585 |
-0.020 |
| 236 |
256334_at |
AT1G72125
|
AtNPF5.13, NPF5.13, NRT1/ PTR family 5.13 |
0.583 |
-0.014 |
| 237 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.583 |
-0.044 |
| 238 |
245663_at |
AT1G28170
|
SOT7, sulphotransferase 7 |
0.582 |
0.006 |
| 239 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.582 |
0.013 |
| 240 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.581 |
0.021 |
| 241 |
253796_at |
AT4G28460
|
unknown |
0.581 |
-0.034 |
| 242 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.580 |
-0.002 |
| 243 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
0.580 |
-0.036 |
| 244 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.579 |
0.124 |
| 245 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
0.579 |
-0.067 |
| 246 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.578 |
0.010 |
| 247 |
245566_at |
AT4G14610
|
[pseudogene] |
0.578 |
-0.072 |
| 248 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
0.577 |
-0.008 |
| 249 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.577 |
-0.026 |
| 250 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.577 |
-0.008 |
| 251 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
0.571 |
-0.003 |
| 252 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.570 |
0.140 |
| 253 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.570 |
-0.095 |
| 254 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.570 |
0.007 |
| 255 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.568 |
0.089 |
| 256 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.567 |
-0.068 |
| 257 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.566 |
-0.043 |
| 258 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.565 |
0.350 |
| 259 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.565 |
0.009 |
| 260 |
246607_at |
AT5G35370
|
[S-locus lectin protein kinase family protein] |
0.565 |
0.016 |
| 261 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.564 |
0.031 |
| 262 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.563 |
0.106 |
| 263 |
247131_at |
AT5G66190
|
ATLFNR1, LEAF FNR 1, FNR1, ferredoxin-NADP(+)-oxidoreductase 1, LFNR1, leaf-type chloroplast-targeted FNR 1 |
0.562 |
-0.022 |
| 264 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
0.560 |
-0.061 |
| 265 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.560 |
-0.194 |
| 266 |
267410_at |
AT2G34920
|
EDA18, embryo sac development arrest 18 |
0.560 |
-0.040 |
| 267 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.560 |
0.482 |
| 268 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.559 |
-0.171 |
| 269 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
0.558 |
-0.103 |
| 270 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
0.558 |
-0.047 |
| 271 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.557 |
0.003 |
| 272 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
0.557 |
-0.007 |
| 273 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.557 |
-0.074 |
| 274 |
267367_at |
AT2G44210
|
unknown |
0.556 |
-0.060 |
| 275 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.556 |
0.040 |
| 276 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.555 |
-0.022 |
| 277 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.554 |
-0.028 |
| 278 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
0.554 |
-0.057 |
| 279 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.553 |
-0.073 |
| 280 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.552 |
0.021 |
| 281 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.551 |
0.073 |
| 282 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
0.550 |
-0.094 |
| 283 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.550 |
0.011 |
| 284 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
0.550 |
0.008 |
| 285 |
267622_at |
AT2G39690
|
unknown |
0.548 |
-0.025 |
| 286 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
0.547 |
-0.056 |
| 287 |
248568_at |
AT5G49760
|
[Leucine-rich repeat protein kinase family protein] |
0.547 |
-0.029 |
| 288 |
250208_at |
AT5G14000
|
anac084, NAC domain containing protein 84, NAC084, NAC domain containing protein 84 |
0.546 |
-0.034 |
| 289 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.545 |
0.057 |
| 290 |
264238_at |
AT1G54740
|
unknown |
0.544 |
-0.018 |
| 291 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.542 |
-0.078 |
| 292 |
251097_at |
AT5G01560
|
LecRK-VI.4, L-type lectin receptor kinase VI.4, LECRKA4.3, lectin receptor kinase a4.3 |
0.541 |
-0.037 |
| 293 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
0.540 |
-0.065 |
| 294 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.539 |
-0.033 |
| 295 |
260213_at |
AT1G74490
|
[Protein kinase superfamily protein] |
0.539 |
-0.036 |
| 296 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.537 |
-0.005 |
| 297 |
248717_at |
AT5G48175
|
unknown |
0.536 |
-0.030 |
| 298 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.535 |
-0.048 |
| 299 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.535 |
0.237 |
| 300 |
253595_at |
AT4G30830
|
unknown |
0.532 |
-0.004 |
| 301 |
250881_at |
AT5G04080
|
unknown |
-1.138 |
-0.118 |
| 302 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-1.104 |
-0.034 |
| 303 |
252938_at |
AT4G39190
|
unknown |
-1.100 |
0.137 |
| 304 |
255016_at |
AT4G10120
|
ATSPS4F |
-1.066 |
-0.198 |
| 305 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
-1.042 |
-0.068 |
| 306 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-1.015 |
-0.010 |
| 307 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-1.003 |
0.027 |
| 308 |
261215_at |
AT1G32970
|
[Subtilisin-like serine endopeptidase family protein] |
-1.002 |
-0.021 |
| 309 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.984 |
-0.011 |
| 310 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
-0.977 |
-0.007 |
| 311 |
248777_at |
AT5G47920
|
unknown |
-0.965 |
0.053 |
| 312 |
266325_at |
AT2G46630
|
unknown |
-0.965 |
-0.043 |
| 313 |
247070_at |
AT5G66815
|
unknown |
-0.963 |
-0.003 |
| 314 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.934 |
0.014 |
| 315 |
263314_at |
AT2G05760
|
[Xanthine/uracil permease family protein] |
-0.919 |
0.017 |
| 316 |
260112_at |
AT1G63310
|
unknown |
-0.911 |
0.034 |
| 317 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.892 |
-0.003 |
| 318 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
-0.890 |
-0.041 |
| 319 |
263673_at |
AT2G04800
|
unknown |
-0.876 |
0.009 |
| 320 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
-0.875 |
-0.096 |
| 321 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.871 |
0.006 |
| 322 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-0.856 |
-0.012 |
| 323 |
254402_at |
AT4G21310
|
unknown |
-0.843 |
-0.020 |
| 324 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
-0.841 |
-0.013 |
| 325 |
251021_at |
AT5G02140
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.841 |
-0.032 |
| 326 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.840 |
0.072 |
| 327 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.830 |
0.060 |
| 328 |
256574_at |
AT3G14780
|
unknown |
-0.829 |
-0.055 |
| 329 |
266209_at |
AT2G27550
|
ATC, centroradialis |
-0.828 |
-0.031 |
| 330 |
259857_at |
AT1G68430
|
unknown |
-0.819 |
0.062 |
| 331 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-0.814 |
-0.002 |
| 332 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.812 |
-0.007 |
| 333 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.807 |
-0.044 |
| 334 |
266425_at |
AT2G41300
|
SSL1, strictosidine synthase-like 1 |
-0.795 |
0.003 |
| 335 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
-0.795 |
0.000 |
| 336 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.793 |
0.047 |
| 337 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.792 |
0.071 |
| 338 |
248172_at |
AT5G54660
|
[HSP20-like chaperones superfamily protein] |
-0.790 |
-0.116 |
| 339 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.787 |
0.084 |
| 340 |
249332_at |
AT5G40980
|
unknown |
-0.787 |
0.016 |
| 341 |
265057_at |
AT1G52140
|
unknown |
-0.787 |
0.003 |
| 342 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.783 |
-0.039 |
| 343 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.774 |
-0.015 |
| 344 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.771 |
0.025 |
| 345 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
-0.767 |
-0.036 |
| 346 |
258017_at |
AT3G19370
|
unknown |
-0.763 |
-0.011 |
| 347 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.763 |
-0.059 |
| 348 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.761 |
-0.054 |
| 349 |
247879_at |
AT5G57770
|
unknown |
-0.756 |
-0.025 |
| 350 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
-0.751 |
-0.044 |
| 351 |
249367_at |
AT5G40630
|
[Ubiquitin-like superfamily protein] |
-0.750 |
-0.048 |
| 352 |
250464_at |
AT5G10040
|
unknown |
-0.743 |
-0.040 |
| 353 |
263545_at |
AT2G21560
|
unknown |
-0.743 |
0.017 |
| 354 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.741 |
0.027 |
| 355 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.740 |
0.009 |
| 356 |
255494_at |
AT4G02710
|
NET1C, Networked 1C |
-0.740 |
-0.001 |
| 357 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
-0.739 |
0.007 |
| 358 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.736 |
-0.125 |
| 359 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.735 |
-0.014 |
| 360 |
265828_at |
AT2G14520
|
unknown |
-0.733 |
0.022 |
| 361 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
-0.730 |
-0.055 |
| 362 |
256450_at |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.723 |
-0.001 |
| 363 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.723 |
-0.034 |
| 364 |
248936_at |
AT5G45710
|
AT-HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, RHA1, ROOT HANDEDNESS 1 |
-0.723 |
0.002 |
| 365 |
251806_at |
AT3G55370
|
OBP3, OBF-binding protein 3 |
-0.721 |
0.053 |
| 366 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
-0.719 |
-0.033 |
| 367 |
245924_at |
AT5G28750
|
[Bacterial sec-independent translocation protein mttA/Hcf106] |
-0.717 |
0.004 |
| 368 |
262873_at |
AT1G64700
|
unknown |
-0.715 |
0.012 |
| 369 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.715 |
0.000 |
| 370 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.713 |
-0.053 |
| 371 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.711 |
0.161 |
| 372 |
248590_at |
AT5G49660
|
XIP1, XYLEM INTERMIXED WITH PHLOEM 1 |
-0.709 |
-0.046 |
| 373 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.707 |
0.142 |
| 374 |
249484_at |
AT5G38970
|
ATBR6OX, BR6OX, BRASSINOSTEROID-6-OXIDASE, BR6OX1, brassinosteroid-6-oxidase 1, CYP85A1 |
-0.696 |
-0.009 |
| 375 |
248731_at |
AT5G48060
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.694 |
-0.053 |
| 376 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.689 |
-0.070 |
| 377 |
257502_at |
AT1G78110
|
unknown |
-0.689 |
0.052 |
| 378 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
-0.688 |
0.041 |
| 379 |
260389_at |
AT1G74055
|
unknown |
-0.685 |
-0.070 |
| 380 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.685 |
0.053 |
| 381 |
250826_at |
AT5G05220
|
unknown |
-0.684 |
0.003 |
| 382 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
-0.679 |
0.087 |
| 383 |
250284_at |
AT5G13290
|
CRN, CORYNE, SOL2, SUPPRESSOR OF LLP1 2 |
-0.678 |
-0.022 |
| 384 |
266560_at |
AT2G23950
|
[Leucine-rich repeat protein kinase family protein] |
-0.677 |
-0.060 |
| 385 |
265719_at |
AT2G03500
|
[Homeodomain-like superfamily protein] |
-0.675 |
-0.063 |
| 386 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.675 |
-0.027 |
| 387 |
262872_at |
AT1G64690
|
BLT, BRANCHLESS TRICHOMES |
-0.671 |
-0.035 |
| 388 |
256436_at |
AT3G11150
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.670 |
-0.007 |
| 389 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.667 |
0.001 |
| 390 |
253556_at |
AT4G31100
|
[wall-associated kinase, putative] |
-0.666 |
-0.025 |
| 391 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.664 |
-0.071 |
| 392 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.660 |
-0.066 |
| 393 |
245202_at |
AT1G67720
|
[Leucine-rich repeat protein kinase family protein] |
-0.659 |
0.013 |
| 394 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.659 |
0.264 |
| 395 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.658 |
-0.017 |
| 396 |
267311_at |
AT2G34670
|
unknown |
-0.658 |
-0.080 |
| 397 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.656 |
0.082 |
| 398 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.651 |
0.014 |
| 399 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.648 |
-0.160 |
| 400 |
266216_at |
AT2G28810
|
[Dof-type zinc finger DNA-binding family protein] |
-0.646 |
-0.094 |
| 401 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
-0.645 |
-0.061 |
| 402 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.645 |
-0.011 |
| 403 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.643 |
-0.134 |
| 404 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.643 |
0.006 |
| 405 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
-0.643 |
0.007 |
| 406 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.640 |
1.047 |
| 407 |
246231_at |
AT4G37080
|
unknown |
-0.640 |
-0.101 |
| 408 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
-0.639 |
-0.076 |
| 409 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
-0.639 |
0.007 |
| 410 |
249574_at |
AT5G37660
|
PDLP7, plasmodesmata-located protein 7 |
-0.636 |
-0.014 |
| 411 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.635 |
-0.027 |
| 412 |
246125_at |
AT5G19875
|
unknown |
-0.634 |
-0.045 |
| 413 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.633 |
-0.015 |
| 414 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
-0.632 |
-0.039 |
| 415 |
265632_at |
AT2G14290
|
unknown |
-0.632 |
0.016 |
| 416 |
247974_at |
AT5G56780
|
ATET2, ARABIDOPSIS EFFECTOR OF TRANSCRIPTION2, ET2, effector of transcription2 |
-0.626 |
-0.053 |
| 417 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.625 |
-0.031 |
| 418 |
258196_at |
AT3G13980
|
unknown |
-0.624 |
0.148 |
| 419 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
-0.622 |
-0.029 |
| 420 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.621 |
-0.161 |
| 421 |
252586_at |
AT3G45610
|
DOF6, DOF transcription factor 6 |
-0.617 |
0.034 |
| 422 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.616 |
0.049 |
| 423 |
254362_at |
AT4G22160
|
unknown |
-0.614 |
0.072 |
| 424 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.610 |
0.055 |
| 425 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.609 |
-0.086 |
| 426 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.608 |
-0.013 |
| 427 |
264718_at |
AT1G70130
|
LecRK-V.2, L-type lectin receptor kinase V.2 |
-0.607 |
-0.017 |
| 428 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
-0.605 |
-0.031 |
| 429 |
249388_at |
AT5G40090
|
CHL1, CHS1-like 1 |
-0.605 |
0.003 |
| 430 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.603 |
0.198 |
| 431 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
-0.602 |
-0.043 |
| 432 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
-0.601 |
-0.031 |
| 433 |
265204_at |
AT2G36650
|
unknown |
-0.601 |
0.025 |
| 434 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.600 |
-0.101 |
| 435 |
245990_at |
AT5G20640
|
unknown |
-0.598 |
-0.032 |
| 436 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
-0.593 |
0.033 |
| 437 |
246833_at |
AT5G26620
|
unknown |
-0.591 |
-0.009 |
| 438 |
265873_at |
AT2G01630
|
[O-Glycosyl hydrolases family 17 protein] |
-0.591 |
0.053 |
| 439 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.590 |
-0.198 |
| 440 |
261684_at |
AT1G47400
|
unknown |
-0.589 |
0.496 |
| 441 |
247716_at |
AT5G59350
|
unknown |
-0.589 |
-0.116 |
| 442 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
-0.587 |
-0.170 |
| 443 |
247897_at |
AT5G57810
|
TET15, tetraspanin15 |
-0.585 |
-0.023 |
| 444 |
264947_at |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.584 |
-0.023 |
| 445 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.584 |
-0.124 |
| 446 |
250348_at |
AT5G11990
|
[proline-rich family protein] |
-0.583 |
-0.001 |
| 447 |
247027_at |
AT5G67090
|
[Subtilisin-like serine endopeptidase family protein] |
-0.581 |
-0.001 |
| 448 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.580 |
-0.102 |
| 449 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.579 |
0.050 |
| 450 |
264055_at |
AT2G28590
|
[Protein kinase superfamily protein] |
-0.579 |
-0.068 |
| 451 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.577 |
0.011 |
| 452 |
247357_at |
AT5G63710
|
[Leucine-rich repeat protein kinase family protein] |
-0.577 |
-0.063 |
| 453 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
-0.576 |
-0.021 |
| 454 |
251481_at |
AT3G59730
|
LecRK-V.6, L-type lectin receptor kinase V.6 |
-0.575 |
-0.003 |
| 455 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.575 |
0.061 |
| 456 |
260341_at |
AT1G69170
|
AtSPL6, SPL6, SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-domain transcription factor 6 |
-0.573 |
-0.096 |
| 457 |
263002_at |
AT1G54200
|
unknown |
-0.572 |
0.101 |
| 458 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.571 |
0.056 |
| 459 |
248535_at |
AT5G50120
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.571 |
0.003 |
| 460 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.571 |
-0.059 |
| 461 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
-0.570 |
-0.061 |
| 462 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
-0.567 |
0.013 |
| 463 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
-0.566 |
-0.033 |
| 464 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
-0.564 |
0.075 |
| 465 |
245087_at |
AT2G39830
|
DAR2, DA1-related protein 2, LRD3, LATERAL ROOT DEVELOPMENT 3 |
-0.562 |
-0.054 |
| 466 |
261345_at |
AT1G79760
|
DTA4, downstream target of AGL15-4 |
-0.560 |
0.023 |
| 467 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
-0.560 |
-0.029 |
| 468 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.558 |
0.050 |
| 469 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
-0.556 |
0.136 |
| 470 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
-0.556 |
0.031 |
| 471 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.555 |
0.029 |
| 472 |
248620_at |
AT5G49320
|
unknown |
-0.555 |
-0.046 |
| 473 |
257956_at |
AT3G25400
|
unknown |
-0.554 |
0.069 |
| 474 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.553 |
-0.135 |
| 475 |
259565_at |
AT1G20530
|
unknown |
-0.553 |
-0.007 |
| 476 |
249256_at |
AT5G41620
|
unknown |
-0.552 |
-0.124 |
| 477 |
262628_at |
AT1G06490
|
ATGSL07, Arabidopsis thaliana glucan synthase-like 7, atgsl7, CalS7, callose synthase 7, gsl07, GSL07, glucan synthase-like 7, GSL7, glucan synthase-like 7 |
-0.549 |
-0.055 |
| 478 |
249246_at |
AT5G42290
|
[transcription activator-related] |
-0.548 |
-0.014 |
| 479 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
-0.547 |
-0.049 |
| 480 |
256568_at |
AT3G19520
|
unknown |
-0.547 |
0.028 |
| 481 |
252341_at |
AT3G48940
|
[Remorin family protein] |
-0.547 |
-0.029 |
| 482 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.547 |
-0.050 |
| 483 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.545 |
-0.031 |
| 484 |
257517_at |
AT3G16330
|
unknown |
-0.541 |
0.030 |
| 485 |
245755_at |
AT1G35210
|
unknown |
-0.541 |
0.035 |
| 486 |
264345_at |
AT1G11915
|
unknown |
-0.540 |
-0.024 |
| 487 |
258828_at |
AT3G07130
|
ATPAP15, PURPLE ACID PHOSPHATASE 15, PAP15, purple acid phosphatase 15 |
-0.539 |
-0.125 |
| 488 |
246842_at |
AT5G26731
|
unknown |
-0.538 |
0.051 |
| 489 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
-0.537 |
0.050 |
| 490 |
256564_at |
AT3G29770
|
ATMES11, ARABIDOPSIS THALIANA METHYL ESTERASE 11, MES11, methyl esterase 11 |
-0.537 |
-0.046 |
| 491 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.536 |
-0.052 |
| 492 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.535 |
0.116 |
| 493 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.535 |
0.018 |
| 494 |
265769_at |
AT2G48090
|
unknown |
-0.535 |
-0.055 |
| 495 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.535 |
-0.108 |
| 496 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.534 |
-0.024 |
| 497 |
265173_at |
AT1G23530
|
unknown |
-0.533 |
-0.059 |
| 498 |
247985_at |
AT5G56790
|
[Protein kinase superfamily protein] |
-0.533 |
-0.047 |
| 499 |
258197_at |
AT3G14000
|
ATBRXL2, BRX-LIKE2 |
-0.530 |
-0.019 |
| 500 |
253074_at |
AT4G36140
|
[disease resistance protein (TIR-NBS-LRR class), putative] |
-0.529 |
-0.028 |
| 501 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
-0.526 |
0.051 |
| 502 |
256734_at |
AT3G29390
|
RIK, RS2-interacting KH protein |
-0.526 |
0.030 |
| 503 |
257511_at |
AT1G43000
|
[PLATZ transcription factor family protein] |
-0.523 |
-0.036 |
| 504 |
249724_at |
AT5G35450
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.523 |
0.020 |
| 505 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.522 |
-0.016 |
| 506 |
248628_at |
AT5G48960
|
[HAD-superfamily hydrolase, subfamily IG, 5'-nucleotidase] |
-0.520 |
0.091 |
| 507 |
255876_at |
AT2G40480
|
unknown |
-0.520 |
0.031 |
| 508 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.520 |
-0.078 |
| 509 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.519 |
-0.168 |
| 510 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
-0.518 |
-0.044 |
| 511 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.514 |
-0.007 |
| 512 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.514 |
-0.018 |
| 513 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.513 |
-0.029 |
| 514 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
-0.510 |
0.066 |
| 515 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
-0.510 |
-0.006 |
| 516 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.509 |
-0.026 |
| 517 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.508 |
0.042 |
| 518 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.508 |
0.041 |
| 519 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
-0.507 |
-0.014 |
| 520 |
251162_at |
AT3G63300
|
FKD1, FORKED 1 |
-0.507 |
-0.022 |
| 521 |
250333_at |
AT5G11690
|
ATTIM17-3, ARABIDOPSIS THALIANA TRANSLOCASE INNER MEMBRANE SUBUNIT 17-3, TIM17-3, translocase inner membrane subunit 17-3 |
-0.506 |
-0.027 |
| 522 |
252072_at |
AT3G51710
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
-0.503 |
0.009 |
| 523 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.502 |
0.065 |
| 524 |
262392_at |
AT1G49520
|
[SWIB complex BAF60b domain-containing protein] |
-0.502 |
-0.027 |
| 525 |
258213_at |
AT3G17950
|
unknown |
-0.501 |
0.035 |
| 526 |
247837_at |
AT5G57840
|
[HXXXD-type acyl-transferase family protein] |
-0.500 |
-0.022 |
| 527 |
246888_at |
AT5G26270
|
unknown |
-0.499 |
-0.040 |
| 528 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.498 |
0.007 |
| 529 |
249034_at |
AT5G44160
|
AtIDD8, IDD8, INDETERMINATE DOMAIN 8, NUC, nutcracker |
-0.498 |
-0.040 |
| 530 |
251290_at |
AT3G61850
|
DAG1, dof affecting germination 1 |
-0.494 |
0.013 |
| 531 |
259752_at |
AT1G71040
|
LPR2, Low Phosphate Root2 |
-0.493 |
-0.005 |
| 532 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.492 |
0.014 |
| 533 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.491 |
-0.024 |
| 534 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.490 |
-0.007 |
| 535 |
264605_at |
AT1G04550
|
BDL, BODENLOS, IAA12, indole-3-acetic acid inducible 12 |
-0.490 |
-0.053 |
| 536 |
250265_at |
AT5G12900
|
unknown |
-0.489 |
0.017 |
| 537 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.489 |
-0.119 |
| 538 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.488 |
0.017 |
| 539 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.488 |
-0.034 |
| 540 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
-0.488 |
0.016 |
| 541 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.486 |
0.087 |
| 542 |
248079_at |
AT5G55790
|
unknown |
-0.486 |
0.028 |
| 543 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.486 |
-0.048 |
| 544 |
248261_at |
AT5G53280
|
PDV1, PLASTID DIVISION1 |
-0.486 |
-0.132 |
| 545 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
-0.485 |
-0.040 |
| 546 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.485 |
0.033 |
| 547 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.485 |
-0.046 |
| 548 |
247948_at |
AT5G57130
|
SMXL5, SMAX1-like 5 |
-0.484 |
0.080 |
| 549 |
247397_at |
AT5G62940
|
DOF5.6, DNA BINDING WITH ONE FINGER 5.6, HCA2, HIGH CAMBIAL ACTIVITY2 |
-0.484 |
0.004 |
| 550 |
246735_at |
AT5G27670
|
HTA7, histone H2A 7 |
-0.483 |
0.048 |
| 551 |
251476_at |
AT3G59670
|
unknown |
-0.482 |
-0.030 |
| 552 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.482 |
0.040 |
| 553 |
259566_at |
AT1G20520
|
unknown |
-0.481 |
-0.025 |
| 554 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
-0.481 |
0.022 |
| 555 |
253516_at |
AT4G31360
|
[selenium binding] |
-0.480 |
0.080 |
| 556 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.480 |
0.103 |
| 557 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.479 |
0.011 |
| 558 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.479 |
0.089 |
| 559 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
-0.479 |
-0.031 |
| 560 |
249821_at |
AT5G23690
|
[Polynucleotide adenylyltransferase family protein] |
-0.478 |
-0.036 |
| 561 |
261646_at |
AT1G27690
|
unknown |
-0.478 |
-0.062 |
| 562 |
249904_at |
AT5G22700
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.477 |
0.067 |
| 563 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.476 |
0.056 |
| 564 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
-0.473 |
-0.074 |
| 565 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
-0.472 |
0.018 |
| 566 |
249622_at |
AT5G37550
|
unknown |
-0.471 |
-0.068 |
| 567 |
266812_at |
AT2G44830
|
[Protein kinase superfamily protein] |
-0.471 |
-0.029 |
| 568 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.471 |
-0.100 |
| 569 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.471 |
0.042 |
| 570 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
-0.470 |
-0.051 |
| 571 |
245758_at |
AT1G32240
|
KAN2, KANADI 2 |
-0.470 |
-0.050 |
| 572 |
258608_at |
AT3G03020
|
unknown |
-0.469 |
0.111 |
| 573 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
-0.468 |
0.013 |
| 574 |
267171_at |
AT2G37590
|
ATDOF2.4, DOF2.4, DNA binding with one finger 2.4 |
-0.467 |
0.015 |
| 575 |
247986_at |
AT5G56880
|
unknown |
-0.467 |
0.004 |
| 576 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
-0.466 |
0.224 |
| 577 |
262345_at |
AT1G64180
|
[intracellular protein transport protein USO1-related] |
-0.465 |
-0.086 |
| 578 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.464 |
-0.102 |
| 579 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.464 |
0.094 |
| 580 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.464 |
-0.065 |
| 581 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.464 |
0.054 |
| 582 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.464 |
-0.066 |
| 583 |
264682_at |
AT1G65570
|
[Pectin lyase-like superfamily protein] |
-0.464 |
-0.051 |
| 584 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.464 |
0.039 |
| 585 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.464 |
0.011 |
| 586 |
249312_at |
AT5G41550
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.463 |
-0.010 |
| 587 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.463 |
0.041 |
| 588 |
252164_at |
AT3G50620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.461 |
0.073 |
| 589 |
265326_at |
AT2G18220
|
[Noc2p family] |
-0.461 |
0.049 |
| 590 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.460 |
0.102 |
| 591 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.460 |
-0.034 |
| 592 |
250472_at |
AT5G10210
|
unknown |
-0.460 |
-0.029 |
| 593 |
264277_at |
AT1G60390
|
PG1, polygalacturonase 1 |
-0.459 |
0.022 |
| 594 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
-0.458 |
0.020 |
| 595 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
-0.458 |
-0.067 |
| 596 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.458 |
-0.192 |
| 597 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.457 |
0.060 |
| 598 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.456 |
-0.022 |
| 599 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.455 |
-0.031 |
| 600 |
247180_at |
AT5G65350
|
HTR11, histone 3 11 |
-0.455 |
0.078 |