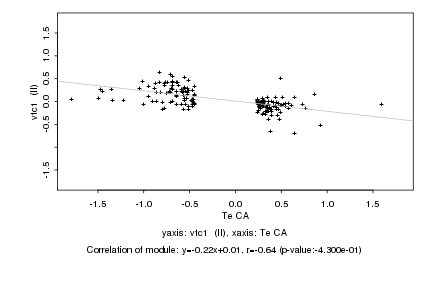
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Te CA |
Log2 signal ratio
vtc1 (II) |
| 1 |
265889_at |
AT2G15130
|
[Plant basic secretory protein (BSP) family protein] |
1.587 |
-0.052 |
| 2 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.927 |
-0.524 |
| 3 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.859 |
0.174 |
| 4 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.756 |
-0.139 |
| 5 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.730 |
-0.048 |
| 6 |
246406_at |
AT1G57650
|
[ATP binding] |
0.644 |
0.101 |
| 7 |
262421_at |
AT1G50290
|
unknown |
0.635 |
-0.684 |
| 8 |
256859_at |
AT3G22940
|
[F-box associated ubiquitination effector family protein] |
0.609 |
-0.066 |
| 9 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.572 |
-0.146 |
| 10 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.569 |
-0.032 |
| 11 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
0.545 |
-0.098 |
| 12 |
247797_at |
AT5G58780
|
AtcPT5, AtHEPS, cPT5, cis -prenyltransferase 5, HEPS, heptaprenyl diphosphate synthase |
0.544 |
-0.025 |
| 13 |
262321_at |
AT1G27570
|
[phosphatidylinositol 3- and 4-kinase family protein] |
0.517 |
-0.048 |
| 14 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.504 |
0.099 |
| 15 |
256175_at |
AT1G51670
|
unknown |
0.494 |
-0.072 |
| 16 |
261302_at |
AT1G48580
|
unknown |
0.492 |
-0.075 |
| 17 |
251344_at |
AT3G60920
|
unknown |
0.485 |
-0.086 |
| 18 |
252345_at |
AT3G48640
|
unknown |
0.483 |
0.508 |
| 19 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.482 |
-0.232 |
| 20 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.474 |
-0.384 |
| 21 |
255653_at |
AT4G00960
|
[Protein kinase superfamily protein] |
0.468 |
-0.023 |
| 22 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.466 |
-0.156 |
| 23 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
0.448 |
-0.300 |
| 24 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.445 |
-0.158 |
| 25 |
246344_at |
AT3G56730
|
[Putative endonuclease or glycosyl hydrolase] |
0.443 |
-0.129 |
| 26 |
261665_at |
AT1G18310
|
[glycosyl hydrolase family 81 protein] |
0.439 |
-0.003 |
| 27 |
248695_at |
AT5G48350
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.436 |
-0.067 |
| 28 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.432 |
0.092 |
| 29 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.407 |
-0.108 |
| 30 |
244901_at |
ATMG00640
|
ORF25 |
0.406 |
-0.004 |
| 31 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
0.393 |
-0.160 |
| 32 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.391 |
-0.293 |
| 33 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.387 |
-0.208 |
| 34 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.387 |
0.021 |
| 35 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.381 |
-0.160 |
| 36 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.378 |
-0.645 |
| 37 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.370 |
-0.118 |
| 38 |
250136_at |
AT5G15380
|
DRM1, domains rearranged methylase 1 |
0.360 |
-0.054 |
| 39 |
266355_at |
AT2G01400
|
unknown |
0.357 |
-0.217 |
| 40 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.354 |
-0.382 |
| 41 |
264104_at |
AT2G13750
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.5e-17 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.351 |
0.014 |
| 42 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.345 |
-0.212 |
| 43 |
244932_at |
ATCG01060
|
PSAC |
0.344 |
-0.086 |
| 44 |
261001_at |
AT1G26530
|
[PIN domain-like family protein] |
0.344 |
-0.134 |
| 45 |
249397_at |
AT5G40230
|
UMAMIT37, Usually multiple acids move in and out Transporters 37 |
0.343 |
0.097 |
| 46 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.328 |
-0.129 |
| 47 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.328 |
-0.206 |
| 48 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.321 |
-0.210 |
| 49 |
245215_at |
AT1G67830
|
ATFXG1, Arabidopsis thaliana alpha-fucosidase 1, FXG1, alpha-fucosidase 1 |
0.318 |
-0.274 |
| 50 |
251345_at |
AT3G60940
|
[Putative endonuclease or glycosyl hydrolase] |
0.308 |
0.010 |
| 51 |
246590_at |
AT5G14870
|
ATCNGC18, CYCLIC NUCLEOTIDE-GATED CHANNEL 18, CNGC18, cyclic nucleotide-gated channel 18 |
0.306 |
-0.052 |
| 52 |
245743_at |
AT1G51080
|
unknown |
0.305 |
-0.260 |
| 53 |
249802_at |
AT5G23770
|
unknown |
0.304 |
-0.106 |
| 54 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.302 |
0.035 |
| 55 |
249107_at |
AT5G43680
|
unknown |
0.302 |
-0.002 |
| 56 |
246757_at |
AT5G27940
|
WPP3, WPP domain protein 3 |
0.301 |
-0.022 |
| 57 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
0.297 |
-0.106 |
| 58 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.295 |
0.079 |
| 59 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.294 |
-0.089 |
| 60 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.294 |
0.020 |
| 61 |
249553_at |
AT5G38260
|
[Protein kinase superfamily protein] |
0.294 |
-0.271 |
| 62 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.293 |
-0.016 |
| 63 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.290 |
-0.100 |
| 64 |
257158_at |
AT3G24360
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.288 |
-0.078 |
| 65 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.287 |
0.035 |
| 66 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.272 |
-0.094 |
| 67 |
249147_at |
AT5G43330
|
c-NAD-MDH2, cytosolic-NAD-dependent malate dehydrogenase 2 |
0.266 |
-0.010 |
| 68 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
0.266 |
-0.035 |
| 69 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
0.266 |
-0.147 |
| 70 |
261277_at |
AT1G20230
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.259 |
-0.081 |
| 71 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
0.256 |
-0.117 |
| 72 |
246316_at |
AT3G56890
|
[F-box associated ubiquitination effector family protein] |
0.254 |
-0.106 |
| 73 |
255760_at |
AT1G16780
|
AtVHP2;2, VHP2;2 |
0.252 |
-0.103 |
| 74 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.250 |
-0.062 |
| 75 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
0.250 |
-0.028 |
| 76 |
249726_at |
AT5G35480
|
unknown |
0.247 |
-0.003 |
| 77 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
0.245 |
0.015 |
| 78 |
251823_at |
AT3G55080
|
[SET domain-containing protein] |
0.245 |
-0.043 |
| 79 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
0.244 |
-0.051 |
| 80 |
252946_at |
AT4G39235
|
unknown |
0.243 |
-0.185 |
| 81 |
253513_at |
AT4G31760
|
[Peroxidase superfamily protein] |
0.239 |
0.029 |
| 82 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.237 |
-0.225 |
| 83 |
259539_at |
AT1G20550
|
[O-fucosyltransferase family protein] |
0.234 |
-0.015 |
| 84 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.234 |
0.001 |
| 85 |
264167_at |
AT1G02060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.234 |
0.058 |
| 86 |
264068_at |
AT2G27990
|
BLH8, BEL1-like homeodomain 8, PNF, POUND-FOOLISH |
0.233 |
0.060 |
| 87 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.796 |
0.038 |
| 88 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-1.494 |
0.085 |
| 89 |
252462_at |
AT3G47250
|
unknown |
-1.479 |
0.269 |
| 90 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-1.450 |
0.223 |
| 91 |
264958_at |
AT1G76960
|
unknown |
-1.362 |
0.279 |
| 92 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-1.347 |
0.038 |
| 93 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.222 |
0.027 |
| 94 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-1.048 |
0.291 |
| 95 |
267345_at |
AT2G44240
|
unknown |
-1.020 |
0.460 |
| 96 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-1.007 |
-0.045 |
| 97 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.957 |
0.341 |
| 98 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.957 |
0.123 |
| 99 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.907 |
0.015 |
| 100 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
-0.887 |
0.303 |
| 101 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
-0.880 |
0.399 |
| 102 |
263023_at |
AT1G23960
|
unknown |
-0.869 |
0.217 |
| 103 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.864 |
0.001 |
| 104 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.835 |
0.423 |
| 105 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.834 |
0.639 |
| 106 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
-0.822 |
0.216 |
| 107 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.799 |
-0.166 |
| 108 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.796 |
-0.019 |
| 109 |
252659_at |
AT3G44430
|
unknown |
-0.783 |
-0.148 |
| 110 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.782 |
0.396 |
| 111 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.777 |
0.358 |
| 112 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.764 |
0.429 |
| 113 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.752 |
0.195 |
| 114 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.743 |
0.422 |
| 115 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.722 |
0.199 |
| 116 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.722 |
0.238 |
| 117 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.718 |
0.216 |
| 118 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.716 |
-0.002 |
| 119 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.710 |
0.602 |
| 120 |
249896_at |
AT5G22530
|
unknown |
-0.705 |
0.304 |
| 121 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.697 |
0.457 |
| 122 |
251633_at |
AT3G57460
|
[catalytics] |
-0.696 |
0.276 |
| 123 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.696 |
0.269 |
| 124 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.695 |
0.550 |
| 125 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-0.693 |
0.371 |
| 126 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.692 |
0.431 |
| 127 |
256617_at |
AT3G22240
|
unknown |
-0.688 |
0.017 |
| 128 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.653 |
0.418 |
| 129 |
263007_at |
AT1G54260
|
[winged-helix DNA-binding transcription factor family protein] |
-0.653 |
0.228 |
| 130 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.648 |
0.131 |
| 131 |
252676_at |
AT3G44280
|
unknown |
-0.646 |
-0.048 |
| 132 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.644 |
0.146 |
| 133 |
254735_at |
AT4G13810
|
AtRLP47, receptor like protein 47, RLP47, receptor like protein 47 |
-0.640 |
0.426 |
| 134 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.625 |
0.356 |
| 135 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.598 |
0.266 |
| 136 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.596 |
0.224 |
| 137 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
-0.590 |
-0.048 |
| 138 |
260110_at |
AT1G63350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.582 |
0.204 |
| 139 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
-0.574 |
0.241 |
| 140 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.571 |
-0.156 |
| 141 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.570 |
0.149 |
| 142 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.563 |
0.325 |
| 143 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.563 |
0.547 |
| 144 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.560 |
0.084 |
| 145 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.559 |
0.040 |
| 146 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.558 |
0.302 |
| 147 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
-0.553 |
0.233 |
| 148 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.549 |
-0.049 |
| 149 |
248079_at |
AT5G55790
|
unknown |
-0.540 |
0.085 |
| 150 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.538 |
0.076 |
| 151 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.529 |
0.154 |
| 152 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.527 |
0.204 |
| 153 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.527 |
0.286 |
| 154 |
258027_at |
AT3G19515
|
unknown |
-0.522 |
-0.108 |
| 155 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.519 |
0.258 |
| 156 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.518 |
-0.155 |
| 157 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
-0.516 |
0.468 |
| 158 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.488 |
0.027 |
| 159 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
-0.477 |
0.260 |
| 160 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.472 |
0.006 |
| 161 |
254594_at |
AT4G18930
|
[RNA ligase/cyclic nucleotide phosphodiesterase family protein] |
-0.471 |
0.055 |
| 162 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.471 |
-0.049 |
| 163 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.469 |
-0.097 |
| 164 |
245453_at |
AT4G16900
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.462 |
-0.019 |
| 165 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.461 |
0.054 |
| 166 |
248936_at |
AT5G45710
|
AT-HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, RHA1, ROOT HANDEDNESS 1 |
-0.458 |
-0.030 |
| 167 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.455 |
-0.039 |
| 168 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.453 |
0.349 |
| 169 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.451 |
-0.059 |
| 170 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
-0.449 |
0.174 |
| 171 |
250740_at |
AT5G05760
|
ATSED5, ATSYP31, SED5, T-SNARE SED 5, SYP31, syntaxin of plants 31 |
-0.447 |
0.150 |