|
probeID |
AGICode |
Annotation |
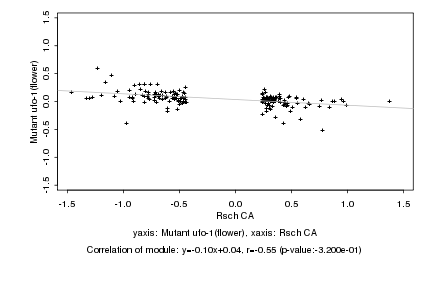
Log2 signal ratio
Rsch CA |
Log2 signal ratio
Mutant ufo-1(flower) |
| 1 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
1.370 |
0.012 |
| 2 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.986 |
-0.065 |
| 3 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.959 |
0.011 |
| 4 |
249259_at |
AT5G41660
|
unknown |
0.945 |
0.042 |
| 5 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.884 |
0.010 |
| 6 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.864 |
0.015 |
| 7 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.834 |
-0.100 |
| 8 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.775 |
-0.505 |
| 9 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.762 |
0.032 |
| 10 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.743 |
-0.073 |
| 11 |
250135_at |
AT5G15360
|
unknown |
0.654 |
-0.039 |
| 12 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.644 |
-0.027 |
| 13 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.625 |
-0.101 |
| 14 |
253689_at |
AT4G29770
|
unknown |
0.604 |
0.051 |
| 15 |
262421_at |
AT1G50290
|
unknown |
0.576 |
-0.307 |
| 16 |
246703_at |
AT5G28080
|
WNK9 |
0.551 |
-0.028 |
| 17 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.545 |
0.085 |
| 18 |
255371_at |
AT4G04110
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.544 |
0.056 |
| 19 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.501 |
-0.098 |
| 20 |
254595_at |
AT4G18960
|
AG, AGAMOUS |
0.489 |
-0.171 |
| 21 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.481 |
0.095 |
| 22 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
0.470 |
0.077 |
| 23 |
257670_at |
AT3G20340
|
unknown |
0.462 |
-0.018 |
| 24 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.461 |
-0.062 |
| 25 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.449 |
-0.075 |
| 26 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.448 |
-0.072 |
| 27 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.444 |
-0.023 |
| 28 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.435 |
-0.006 |
| 29 |
263674_at |
AT2G04790
|
unknown |
0.432 |
0.031 |
| 30 |
246757_at |
AT5G27940
|
WPP3, WPP domain protein 3 |
0.431 |
-0.043 |
| 31 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.423 |
-0.055 |
| 32 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.423 |
-0.382 |
| 33 |
245215_at |
AT1G67830
|
ATFXG1, Arabidopsis thaliana alpha-fucosidase 1, FXG1, alpha-fucosidase 1 |
0.401 |
0.060 |
| 34 |
252744_at |
AT3G43310
|
[pseudogene] |
0.400 |
-0.004 |
| 35 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.390 |
0.099 |
| 36 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.387 |
0.055 |
| 37 |
256175_at |
AT1G51670
|
unknown |
0.386 |
0.131 |
| 38 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
0.379 |
0.015 |
| 39 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
0.377 |
0.015 |
| 40 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.366 |
0.079 |
| 41 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.357 |
-0.274 |
| 42 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.356 |
0.085 |
| 43 |
246793_at |
AT5G27210
|
Cand8, candidate G-protein Coupled Receptor 8 |
0.350 |
0.005 |
| 44 |
259049_at |
AT3G03370
|
unknown |
0.343 |
0.042 |
| 45 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.343 |
0.028 |
| 46 |
266355_at |
AT2G01400
|
unknown |
0.342 |
0.042 |
| 47 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.337 |
0.079 |
| 48 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.334 |
-0.015 |
| 49 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.326 |
0.045 |
| 50 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.325 |
-0.074 |
| 51 |
248425_at |
AT5G51690
|
ACS12, 1-amino-cyclopropane-1-carboxylate synthase 12 |
0.317 |
0.010 |
| 52 |
248969_at |
AT5G45310
|
unknown |
0.316 |
0.066 |
| 53 |
254612_at |
AT4G19100
|
PAM68, photosynthesis affected mutant 68 |
0.315 |
0.088 |
| 54 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.311 |
-0.126 |
| 55 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
0.306 |
0.001 |
| 56 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.305 |
0.105 |
| 57 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.302 |
-0.067 |
| 58 |
245446_at |
AT4G16800
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.301 |
0.035 |
| 59 |
263151_at |
AT1G54120
|
unknown |
0.300 |
-0.113 |
| 60 |
258191_at |
AT3G29100
|
ATVTI13, VTI13, vesicle transport V-snare 13 |
0.296 |
0.071 |
| 61 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.294 |
0.053 |
| 62 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
0.293 |
-0.020 |
| 63 |
264761_at |
AT1G61280
|
[Phosphatidylinositol N-acetylglucosaminyltransferase, GPI19/PIG-P subunit] |
0.289 |
0.027 |
| 64 |
259728_at |
AT1G60970
|
[SNARE-like superfamily protein] |
0.289 |
0.033 |
| 65 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.283 |
0.059 |
| 66 |
252036_at |
AT3G52070
|
unknown |
0.283 |
0.084 |
| 67 |
249995_at |
AT5G18610
|
[Protein kinase superfamily protein] |
0.278 |
0.028 |
| 68 |
257158_at |
AT3G24360
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.278 |
-0.066 |
| 69 |
260381_at |
AT1G73860
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.277 |
-0.175 |
| 70 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.276 |
-0.108 |
| 71 |
253585_at |
AT4G30720
|
PDE327, PIGMENT DEFECTIVE 327 |
0.276 |
0.081 |
| 72 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.273 |
0.081 |
| 73 |
262510_at |
AT1G11270
|
[F-box and associated interaction domains-containing protein] |
0.270 |
0.066 |
| 74 |
254910_at |
AT4G11175
|
[Nucleic acid-binding, OB-fold-like protein] |
0.270 |
0.024 |
| 75 |
264055_at |
AT2G28590
|
[Protein kinase superfamily protein] |
0.268 |
-0.025 |
| 76 |
254529_at |
AT4G19540
|
INDH, iron-sulfur protein required for NADH dehydrogenase, INDL, IND1(iron-sulfur protein required for NADH dehydrogenase)-like |
0.267 |
0.022 |
| 77 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.265 |
0.165 |
| 78 |
259881_at |
AT1G76820
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
0.255 |
0.070 |
| 79 |
249726_at |
AT5G35480
|
unknown |
0.254 |
0.039 |
| 80 |
267110_at |
AT2G14800
|
unknown |
0.252 |
0.054 |
| 81 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.251 |
0.231 |
| 82 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.246 |
0.030 |
| 83 |
267554_at |
AT2G32790
|
[Ubiquitin-conjugating enzyme family protein] |
0.245 |
0.073 |
| 84 |
259030_at |
AT3G09310
|
unknown |
0.242 |
0.009 |
| 85 |
265481_at |
AT2G15960
|
unknown |
0.241 |
0.141 |
| 86 |
266402_at |
AT2G38780
|
unknown |
0.240 |
0.016 |
| 87 |
264431_at |
AT1G61700
|
[RNA polymerases N / 8 kDa subunit] |
0.239 |
-0.222 |
| 88 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
0.238 |
0.145 |
| 89 |
256748_x_at |
AT3G30396
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.1e-39 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.235 |
-0.014 |
| 90 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.234 |
0.040 |
| 91 |
252462_at |
AT3G47250
|
unknown |
-1.470 |
0.164 |
| 92 |
265611_at |
AT2G25510
|
unknown |
-1.335 |
0.069 |
| 93 |
253422_at |
AT4G32240
|
unknown |
-1.305 |
0.061 |
| 94 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-1.277 |
0.073 |
| 95 |
264958_at |
AT1G76960
|
unknown |
-1.239 |
0.603 |
| 96 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.199 |
0.122 |
| 97 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-1.167 |
0.345 |
| 98 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-1.112 |
0.472 |
| 99 |
265831_at |
AT2G14460
|
unknown |
-1.081 |
0.091 |
| 100 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-1.056 |
0.185 |
| 101 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.030 |
0.008 |
| 102 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.977 |
-0.383 |
| 103 |
267345_at |
AT2G44240
|
unknown |
-0.949 |
0.073 |
| 104 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.949 |
0.199 |
| 105 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
-0.925 |
0.079 |
| 106 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.922 |
0.056 |
| 107 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.915 |
0.005 |
| 108 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.902 |
0.303 |
| 109 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.901 |
0.142 |
| 110 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.863 |
0.308 |
| 111 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-0.853 |
0.217 |
| 112 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.835 |
0.113 |
| 113 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.819 |
0.093 |
| 114 |
263023_at |
AT1G23960
|
unknown |
-0.815 |
-0.002 |
| 115 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
-0.814 |
0.321 |
| 116 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.813 |
0.308 |
| 117 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.803 |
0.187 |
| 118 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.793 |
0.106 |
| 119 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.783 |
0.127 |
| 120 |
255627_at |
AT4G00955
|
unknown |
-0.782 |
0.059 |
| 121 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.782 |
0.162 |
| 122 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.768 |
0.043 |
| 123 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.765 |
0.314 |
| 124 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.725 |
0.022 |
| 125 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.724 |
0.087 |
| 126 |
256617_at |
AT3G22240
|
unknown |
-0.723 |
0.132 |
| 127 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.719 |
0.123 |
| 128 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.716 |
0.168 |
| 129 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.713 |
0.137 |
| 130 |
252659_at |
AT3G44430
|
unknown |
-0.713 |
-0.006 |
| 131 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.704 |
0.309 |
| 132 |
249896_at |
AT5G22530
|
unknown |
-0.695 |
0.127 |
| 133 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.691 |
0.106 |
| 134 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.683 |
0.067 |
| 135 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.673 |
0.116 |
| 136 |
251633_at |
AT3G57460
|
[catalytics] |
-0.665 |
0.197 |
| 137 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.659 |
0.119 |
| 138 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
-0.652 |
0.038 |
| 139 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.630 |
0.171 |
| 140 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.627 |
0.072 |
| 141 |
254735_at |
AT4G13810
|
AtRLP47, receptor like protein 47, RLP47, receptor like protein 47 |
-0.624 |
0.079 |
| 142 |
248079_at |
AT5G55790
|
unknown |
-0.619 |
0.105 |
| 143 |
256021_at |
AT1G58270
|
ZW9 |
-0.613 |
-0.177 |
| 144 |
256603_at |
AT3G28270
|
unknown |
-0.611 |
-0.114 |
| 145 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.594 |
0.010 |
| 146 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.583 |
0.173 |
| 147 |
248016_at |
AT5G56380
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.568 |
0.092 |
| 148 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.563 |
0.055 |
| 149 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.556 |
0.068 |
| 150 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.554 |
0.182 |
| 151 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.552 |
0.133 |
| 152 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.546 |
0.040 |
| 153 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.546 |
0.081 |
| 154 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
-0.544 |
0.158 |
| 155 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.535 |
0.094 |
| 156 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.534 |
0.138 |
| 157 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.529 |
0.066 |
| 158 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.522 |
-0.130 |
| 159 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.519 |
0.140 |
| 160 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.517 |
0.033 |
| 161 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.512 |
0.003 |
| 162 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
-0.504 |
0.202 |
| 163 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.500 |
-0.037 |
| 164 |
260110_at |
AT1G63350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.495 |
0.008 |
| 165 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.492 |
0.068 |
| 166 |
251076_at |
AT5G01970
|
unknown |
-0.483 |
0.059 |
| 167 |
249486_at |
AT5G39030
|
[Protein kinase superfamily protein] |
-0.480 |
0.093 |
| 168 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.474 |
-0.031 |
| 169 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.472 |
0.065 |
| 170 |
258424_at |
AT3G16750
|
unknown |
-0.471 |
0.170 |
| 171 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
-0.471 |
-0.012 |
| 172 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
-0.471 |
0.012 |
| 173 |
264666_at |
AT1G09680
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.465 |
0.070 |
| 174 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.461 |
0.161 |
| 175 |
248829_at |
AT5G47130
|
[Bax inhibitor-1 family protein] |
-0.454 |
-0.002 |
| 176 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.454 |
0.261 |
| 177 |
245487_at |
AT4G16250
|
PHYD, phytochrome D |
-0.449 |
0.086 |
| 178 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
-0.448 |
0.039 |
| 179 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
-0.443 |
0.000 |