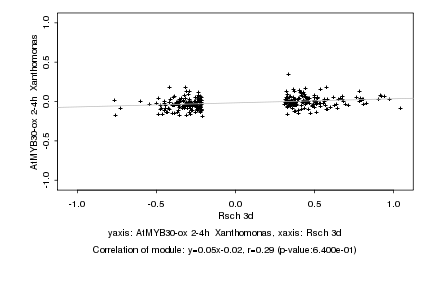
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Rsch 3d |
Log2 signal ratio
AtMYB30-ox 2-4h Xanthomonas |
| 1 |
260668_at |
AT1G19530
|
unknown |
1.042 |
-0.080 |
| 2 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.970 |
0.038 |
| 3 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.941 |
0.076 |
| 4 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.923 |
0.074 |
| 5 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.913 |
0.085 |
| 6 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.903 |
0.031 |
| 7 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.827 |
-0.019 |
| 8 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.810 |
0.029 |
| 9 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.809 |
-0.036 |
| 10 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.799 |
0.035 |
| 11 |
266875_at |
AT2G44800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.797 |
0.015 |
| 12 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.791 |
0.050 |
| 13 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
0.784 |
0.007 |
| 14 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.779 |
0.140 |
| 15 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.765 |
0.056 |
| 16 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.714 |
-0.046 |
| 17 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.693 |
-0.035 |
| 18 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.680 |
0.010 |
| 19 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.677 |
0.071 |
| 20 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.672 |
0.027 |
| 21 |
247535_at |
AT5G61620
|
[myb-like transcription factor family protein] |
0.664 |
0.044 |
| 22 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
0.648 |
0.031 |
| 23 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.644 |
-0.081 |
| 24 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.634 |
-0.030 |
| 25 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
0.625 |
-0.043 |
| 26 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.616 |
0.063 |
| 27 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.600 |
-0.065 |
| 28 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.581 |
0.036 |
| 29 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.580 |
-0.092 |
| 30 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.572 |
0.187 |
| 31 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.572 |
-0.094 |
| 32 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.568 |
-0.050 |
| 33 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
0.561 |
0.037 |
| 34 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.561 |
0.012 |
| 35 |
248801_at |
AT5G47370
|
HAT2 |
0.552 |
-0.022 |
| 36 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.534 |
-0.022 |
| 37 |
259946_at |
AT1G71450
|
[Integrase-type DNA-binding superfamily protein] |
0.534 |
-0.062 |
| 38 |
254468_at |
AT4G20460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.533 |
0.154 |
| 39 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.528 |
-0.023 |
| 40 |
256177_at |
AT1G51620
|
[Protein kinase superfamily protein] |
0.519 |
-0.018 |
| 41 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.519 |
-0.020 |
| 42 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.516 |
0.060 |
| 43 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
0.514 |
0.062 |
| 44 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.514 |
0.043 |
| 45 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
0.511 |
-0.136 |
| 46 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.510 |
-0.021 |
| 47 |
266808_at |
AT2G29995
|
unknown |
0.510 |
0.004 |
| 48 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
0.507 |
-0.028 |
| 49 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.506 |
-0.009 |
| 50 |
250909_at |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.497 |
0.062 |
| 51 |
248141_at |
AT5G55010
|
unknown |
0.497 |
0.009 |
| 52 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.495 |
0.078 |
| 53 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.495 |
-0.072 |
| 54 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.494 |
0.021 |
| 55 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.490 |
-0.034 |
| 56 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.485 |
-0.057 |
| 57 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.484 |
-0.050 |
| 58 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.471 |
-0.152 |
| 59 |
245636_at |
AT1G25240
|
[ENTH/VHS/GAT family protein] |
0.468 |
-0.025 |
| 60 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.466 |
0.040 |
| 61 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.459 |
-0.097 |
| 62 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.459 |
0.040 |
| 63 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.458 |
0.010 |
| 64 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.453 |
0.051 |
| 65 |
258016_at |
AT3G19350
|
MPC, maternally expressed pab C-terminal |
0.449 |
-0.016 |
| 66 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.447 |
0.023 |
| 67 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.446 |
-0.003 |
| 68 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.443 |
0.067 |
| 69 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.442 |
-0.029 |
| 70 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.442 |
0.008 |
| 71 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.440 |
0.176 |
| 72 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.438 |
0.059 |
| 73 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.435 |
0.004 |
| 74 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
0.433 |
0.090 |
| 75 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.432 |
-0.087 |
| 76 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.430 |
-0.003 |
| 77 |
264457_at |
AT1G10400
|
[UDP-Glycosyltransferase superfamily protein] |
0.430 |
0.122 |
| 78 |
254159_at |
AT4G24240
|
ATWRKY7, WRKY7, WRKY DNA-binding protein 7 |
0.429 |
0.001 |
| 79 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.428 |
0.020 |
| 80 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.423 |
-0.001 |
| 81 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.421 |
0.071 |
| 82 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.417 |
0.086 |
| 83 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.415 |
0.127 |
| 84 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
0.415 |
-0.100 |
| 85 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.413 |
0.111 |
| 86 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
0.413 |
-0.049 |
| 87 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.409 |
0.125 |
| 88 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
0.405 |
0.143 |
| 89 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.403 |
0.050 |
| 90 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.399 |
0.049 |
| 91 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.399 |
0.037 |
| 92 |
251822_at |
AT3G55060
|
unknown |
0.399 |
-0.144 |
| 93 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
0.398 |
-0.111 |
| 94 |
253506_at |
AT4G31980
|
unknown |
0.391 |
0.010 |
| 95 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
0.390 |
0.012 |
| 96 |
261085_at |
AT1G17480
|
IQD7, IQ-domain 7 |
0.384 |
-0.040 |
| 97 |
249187_at |
AT5G43060
|
RD21B, esponsive to dehydration 21B |
0.381 |
-0.035 |
| 98 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.379 |
-0.042 |
| 99 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.378 |
-0.020 |
| 100 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.376 |
-0.125 |
| 101 |
258557_at |
AT3G05990
|
[Leucine-rich repeat (LRR) family protein] |
0.376 |
-0.005 |
| 102 |
249068_at |
AT5G43980
|
PDLP1, plasmodesmata-located protein 1, PDLP1A, PLASMODESMATA-LOCATED PROTEIN 1A |
0.374 |
0.033 |
| 103 |
253047_at |
AT4G37295
|
unknown |
0.374 |
-0.034 |
| 104 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.372 |
0.005 |
| 105 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.369 |
-0.119 |
| 106 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
0.369 |
-0.039 |
| 107 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
0.368 |
0.028 |
| 108 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.368 |
0.130 |
| 109 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.366 |
0.156 |
| 110 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
0.365 |
-0.044 |
| 111 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.363 |
-0.001 |
| 112 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.362 |
0.063 |
| 113 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
0.360 |
-0.030 |
| 114 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
0.359 |
-0.034 |
| 115 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.358 |
-0.085 |
| 116 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
0.357 |
-0.022 |
| 117 |
262503_at |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
0.357 |
0.004 |
| 118 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.355 |
0.004 |
| 119 |
262719_at |
AT1G43590
|
unknown |
0.350 |
0.014 |
| 120 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
0.349 |
-0.033 |
| 121 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
0.348 |
-0.025 |
| 122 |
260303_at |
AT1G70520
|
ASG6, ALTERED SEED GERMINATION 6, CRK2, cysteine-rich RLK (RECEPTOR-like protein kinase) 2 |
0.348 |
0.072 |
| 123 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.345 |
-0.002 |
| 124 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
0.345 |
-0.053 |
| 125 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.344 |
0.004 |
| 126 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.342 |
-0.048 |
| 127 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.340 |
-0.005 |
| 128 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
0.340 |
0.009 |
| 129 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.339 |
0.057 |
| 130 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.338 |
0.055 |
| 131 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.337 |
-0.028 |
| 132 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.335 |
0.012 |
| 133 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.334 |
0.346 |
| 134 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.330 |
-0.046 |
| 135 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.328 |
0.046 |
| 136 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.325 |
0.055 |
| 137 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.325 |
-0.075 |
| 138 |
261525_at |
AT1G14330
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.324 |
0.099 |
| 139 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.324 |
-0.058 |
| 140 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
0.324 |
-0.154 |
| 141 |
247726_at |
AT5G59430
|
ATTRP1, TELOMERE REPEAT BINDING PROTEIN 1, TRP1, telomeric repeat binding protein 1 |
0.323 |
0.002 |
| 142 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.322 |
0.135 |
| 143 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.321 |
-0.020 |
| 144 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
0.320 |
0.021 |
| 145 |
256799_at |
AT3G18560
|
unknown |
0.317 |
-0.000 |
| 146 |
259611_at |
AT1G52280
|
AtRABG3d, RAB GTPase homolog G3D, RABG3d, RAB GTPase homolog G3D |
0.317 |
0.059 |
| 147 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.314 |
0.028 |
| 148 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.312 |
-0.001 |
| 149 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.310 |
-0.036 |
| 150 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.769 |
0.015 |
| 151 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.764 |
-0.165 |
| 152 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.728 |
-0.082 |
| 153 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.604 |
0.007 |
| 154 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.544 |
-0.033 |
| 155 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.504 |
-0.019 |
| 156 |
259850_at |
AT1G72240
|
unknown |
-0.492 |
0.043 |
| 157 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.489 |
-0.161 |
| 158 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.478 |
-0.095 |
| 159 |
267063_at |
AT2G41120
|
unknown |
-0.476 |
-0.046 |
| 160 |
252073_at |
AT3G51750
|
unknown |
-0.468 |
-0.081 |
| 161 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.466 |
-0.154 |
| 162 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.454 |
-0.107 |
| 163 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.453 |
0.005 |
| 164 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
-0.452 |
-0.019 |
| 165 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.444 |
-0.038 |
| 166 |
263674_at |
AT2G04790
|
unknown |
-0.444 |
-0.083 |
| 167 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.442 |
-0.134 |
| 168 |
263915_at |
AT2G36430
|
unknown |
-0.433 |
-0.134 |
| 169 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.424 |
-0.088 |
| 170 |
247977_at |
AT5G56850
|
unknown |
-0.422 |
-0.090 |
| 171 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.421 |
-0.010 |
| 172 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
-0.418 |
-0.004 |
| 173 |
256789_at |
AT3G13672
|
SINA2 |
-0.418 |
0.181 |
| 174 |
266781_at |
AT2G28940
|
[Protein kinase superfamily protein] |
-0.411 |
0.037 |
| 175 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.400 |
-0.027 |
| 176 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.399 |
-0.148 |
| 177 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.399 |
-0.046 |
| 178 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.397 |
-0.049 |
| 179 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.393 |
0.055 |
| 180 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.390 |
-0.040 |
| 181 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.389 |
-0.145 |
| 182 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.386 |
0.066 |
| 183 |
245776_at |
AT1G30260
|
unknown |
-0.377 |
-0.029 |
| 184 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.375 |
-0.023 |
| 185 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.374 |
-0.010 |
| 186 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.370 |
-0.130 |
| 187 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.367 |
-0.004 |
| 188 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.367 |
-0.035 |
| 189 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.361 |
-0.039 |
| 190 |
255763_at |
AT1G16730
|
unknown |
-0.361 |
-0.001 |
| 191 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.361 |
-0.103 |
| 192 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.359 |
0.033 |
| 193 |
263987_at |
AT2G42690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.357 |
-0.072 |
| 194 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.356 |
-0.036 |
| 195 |
246330_at |
AT3G43600
|
AAO2, aldehyde oxidase 2, AO3, aldehyde oxidase 3, AOgamma, Aldehyde oxidase gamma, atAO-2, Arabidopsis thaliana aldehyde oxidase 2, AtAO3, Arabidopsis thaliana aldehyde oxidase 3 |
-0.354 |
-0.171 |
| 196 |
246727_at |
AT5G28010
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.354 |
-0.006 |
| 197 |
246576_at |
AT1G31650
|
ATROPGEF14, ROPGEF14, ROP (rho of plants) guanine nucleotide exchange factor 14 |
-0.353 |
0.003 |
| 198 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.345 |
0.042 |
| 199 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.338 |
-0.084 |
| 200 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-0.338 |
-0.053 |
| 201 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.335 |
0.013 |
| 202 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.331 |
-0.055 |
| 203 |
265083_at |
AT1G03820
|
unknown |
-0.329 |
0.008 |
| 204 |
249932_at |
AT5G22390
|
unknown |
-0.328 |
0.081 |
| 205 |
251644_at |
AT3G57540
|
[Remorin family protein] |
-0.328 |
-0.080 |
| 206 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.328 |
-0.060 |
| 207 |
249482_at |
AT5G38980
|
unknown |
-0.325 |
-0.041 |
| 208 |
257966_at |
AT3G19800
|
unknown |
-0.321 |
-0.058 |
| 209 |
262996_at |
AT1G54440
|
[Polynucleotidyl transferase, ribonuclease H fold protein with HRDC domain] |
-0.318 |
0.039 |
| 210 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.317 |
-0.032 |
| 211 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.317 |
0.183 |
| 212 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.316 |
-0.003 |
| 213 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.316 |
0.135 |
| 214 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.312 |
-0.172 |
| 215 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.304 |
-0.014 |
| 216 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.304 |
-0.045 |
| 217 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.304 |
-0.082 |
| 218 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
-0.301 |
0.096 |
| 219 |
248848_at |
AT5G46520
|
VICTR, VARIATION IN COMPOUND TRIGGERED ROOT growth response |
-0.300 |
-0.002 |
| 220 |
262370_at |
AT1G73090
|
unknown |
-0.296 |
-0.057 |
| 221 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.296 |
-0.010 |
| 222 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.295 |
-0.033 |
| 223 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
-0.294 |
-0.038 |
| 224 |
245353_at |
AT4G16000
|
unknown |
-0.293 |
0.129 |
| 225 |
260922_at |
AT1G21560
|
unknown |
-0.292 |
-0.127 |
| 226 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.292 |
-0.029 |
| 227 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
-0.292 |
0.000 |
| 228 |
259207_at |
AT3G09050
|
unknown |
-0.292 |
-0.030 |
| 229 |
254320_at |
AT4G22580
|
[Exostosin family protein] |
-0.291 |
0.036 |
| 230 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.290 |
-0.159 |
| 231 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.286 |
-0.004 |
| 232 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.286 |
-0.047 |
| 233 |
255793_at |
AT2G33250
|
unknown |
-0.285 |
-0.087 |
| 234 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.284 |
-0.110 |
| 235 |
255857_at |
AT1G67080
|
ABA4, abscisic acid (ABA)-deficient 4 |
-0.281 |
-0.031 |
| 236 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.279 |
-0.006 |
| 237 |
262879_at |
AT1G64860
|
RPOD1, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, sigma factor A, SIGB |
-0.279 |
-0.049 |
| 238 |
249579_at |
AT5G37680
|
ARLA1A, ADP-ribosylation factor-like A1A, ATARLA1A, ADP-ribosylation factor-like A1A |
-0.277 |
-0.058 |
| 239 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.277 |
-0.062 |
| 240 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.276 |
-0.113 |
| 241 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.275 |
-0.027 |
| 242 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.271 |
-0.050 |
| 243 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
-0.264 |
0.003 |
| 244 |
250317_at |
AT5G12250
|
TUB6, beta-6 tubulin |
-0.261 |
-0.019 |
| 245 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
-0.258 |
-0.053 |
| 246 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.256 |
0.060 |
| 247 |
263344_at |
AT2G04940
|
[scramblase-related] |
-0.255 |
-0.033 |
| 248 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.254 |
-0.041 |
| 249 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
-0.254 |
0.008 |
| 250 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.253 |
0.006 |
| 251 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.253 |
-0.128 |
| 252 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.253 |
-0.023 |
| 253 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.252 |
-0.053 |
| 254 |
258389_at |
AT3G15390
|
SDE5, silencing defective 5 |
-0.251 |
-0.047 |
| 255 |
254194_at |
AT4G23980
|
ARF9, auxin response factor 9 |
-0.250 |
-0.067 |
| 256 |
250482_at |
AT5G10320
|
unknown |
-0.246 |
0.060 |
| 257 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.244 |
-0.109 |
| 258 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.244 |
-0.101 |
| 259 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
-0.243 |
-0.035 |
| 260 |
255628_at |
AT4G00850
|
GIF3, GRF1-interacting factor 3 |
-0.243 |
0.005 |
| 261 |
251727_at |
AT3G56290
|
unknown |
-0.242 |
0.002 |
| 262 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
-0.242 |
-0.064 |
| 263 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.242 |
-0.008 |
| 264 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.241 |
-0.096 |
| 265 |
250487_at |
AT5G09690
|
ATMGT7, ARABIDOPSIS THALIANA MAGNESIUM TRANSPORTER 7, MGT7, magnesium transporter 7, MRS2-7 |
-0.241 |
-0.012 |
| 266 |
246233_at |
AT4G36550
|
[ARM repeat superfamily protein] |
-0.240 |
0.036 |
| 267 |
257406_at |
AT1G27060
|
[Regulator of chromosome condensation (RCC1) family protein] |
-0.239 |
-0.068 |
| 268 |
259034_at |
AT3G09410
|
[Pectinacetylesterase family protein] |
-0.238 |
0.044 |
| 269 |
260696_at |
AT1G32520
|
unknown |
-0.238 |
-0.081 |
| 270 |
262713_at |
AT1G16520
|
unknown |
-0.235 |
0.122 |
| 271 |
261538_at |
AT1G01830
|
[ARM repeat superfamily protein] |
-0.235 |
0.089 |
| 272 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
-0.234 |
-0.084 |
| 273 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.233 |
-0.060 |
| 274 |
261044_at |
AT1G01290
|
CNX3, cofactor of nitrate reductase and xanthine dehydrogenase 3 |
-0.230 |
0.026 |
| 275 |
249810_at |
AT5G23920
|
unknown |
-0.228 |
-0.001 |
| 276 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.227 |
-0.066 |
| 277 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.227 |
-0.134 |
| 278 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.226 |
0.068 |
| 279 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.226 |
0.020 |
| 280 |
263410_at |
AT2G04039
|
unknown |
-0.225 |
-0.064 |
| 281 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.224 |
-0.125 |
| 282 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.224 |
0.014 |
| 283 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.224 |
-0.049 |
| 284 |
246762_at |
AT5G27620
|
CYCH;1, cyclin H;1 |
-0.223 |
0.031 |
| 285 |
253249_at |
AT4G34680
|
GATA3, GATA transcription factor 3 |
-0.223 |
-0.069 |
| 286 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.222 |
-0.053 |
| 287 |
262604_at |
AT1G15060
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.222 |
-0.000 |
| 288 |
252023_at |
AT3G52920
|
unknown |
-0.221 |
-0.100 |
| 289 |
259505_at |
AT1G15810
|
[S15/NS1, RNA-binding protein] |
-0.221 |
0.060 |
| 290 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.221 |
0.015 |
| 291 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.220 |
-0.098 |
| 292 |
260373_at |
AT1G73970
|
unknown |
-0.219 |
0.000 |
| 293 |
258587_at |
AT3G04310
|
unknown |
-0.218 |
-0.036 |
| 294 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
-0.217 |
-0.103 |
| 295 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
-0.215 |
-0.103 |
| 296 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.215 |
0.013 |
| 297 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.214 |
-0.187 |