|
probeID |
AGICode |
Annotation |
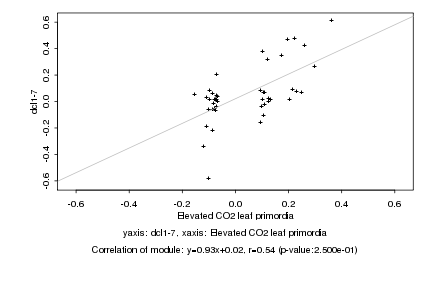
Log2 signal ratio
Elevated CO2 leaf primordia |
Log2 signal ratio
dcl1-7 |
| 1 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.361 |
0.620 |
| 2 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.295 |
0.269 |
| 3 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.259 |
0.429 |
| 4 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.245 |
0.073 |
| 5 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
0.228 |
0.076 |
| 6 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.222 |
0.477 |
| 7 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.212 |
0.096 |
| 8 |
246001_at |
AT5G20790
|
unknown |
0.202 |
0.021 |
| 9 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.194 |
0.472 |
| 10 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.172 |
0.351 |
| 11 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.131 |
0.016 |
| 12 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.122 |
0.025 |
| 13 |
246116_at |
AT5G20310
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.121 |
0.002 |
| 14 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.119 |
0.321 |
| 15 |
251430_at |
AT3G60110
|
[DNA-binding bromodomain-containing protein] |
0.107 |
-0.019 |
| 16 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.106 |
0.071 |
| 17 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.105 |
-0.100 |
| 18 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.103 |
0.075 |
| 19 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
0.100 |
0.385 |
| 20 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.099 |
0.016 |
| 21 |
265878_at |
AT2G42410
|
ATZFP11, ZINC FINGER PROTEIN 11, ZFP11, zinc finger protein 11 |
0.098 |
-0.037 |
| 22 |
252936_at |
AT4G39160
|
[Homeodomain-like superfamily protein] |
0.094 |
0.090 |
| 23 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.094 |
-0.158 |
| 24 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.156 |
0.057 |
| 25 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.122 |
-0.333 |
| 26 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
-0.112 |
-0.184 |
| 27 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.110 |
0.033 |
| 28 |
264151_at |
AT1G02070
|
unknown |
-0.105 |
-0.057 |
| 29 |
256307_at |
AT1G30350
|
[Pectin lyase-like superfamily protein] |
-0.105 |
-0.579 |
| 30 |
259590_at |
AT1G28160
|
[Integrase-type DNA-binding superfamily protein] |
-0.100 |
0.088 |
| 31 |
254895_at |
AT4G11870
|
unknown |
-0.098 |
0.021 |
| 32 |
246182_at |
AT5G20870
|
[O-Glycosyl hydrolases family 17 protein] |
-0.090 |
0.067 |
| 33 |
260131_at |
AT1G66310
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.089 |
-0.059 |
| 34 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.088 |
-0.214 |
| 35 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
-0.083 |
-0.008 |
| 36 |
254302_at |
AT4G22800
|
unknown |
-0.081 |
0.016 |
| 37 |
247887_at |
AT5G57880
|
ATPRD2, ARABIDOPSIS THALIANA PUTATIVE RECOMBINATION INITIATION DEFECTS 2, MPS1, MULTIPOLAR SPINDLE 1, PRD2, PUTATIVE RECOMBINATION INITIATION DEFECTS 2 |
-0.080 |
-0.051 |
| 38 |
250722_at |
AT5G06190
|
unknown |
-0.077 |
-0.062 |
| 39 |
263468_at |
AT2G31930
|
unknown |
-0.076 |
0.019 |
| 40 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
-0.074 |
0.206 |
| 41 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-0.073 |
0.050 |
| 42 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.073 |
-0.033 |
| 43 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
-0.073 |
0.017 |
| 44 |
261278_at |
AT1G05800
|
DGL, DONGLE |
-0.070 |
0.045 |
| 45 |
256357_at |
AT1G66490
|
[F-box and associated interaction domains-containing protein] |
-0.070 |
0.006 |