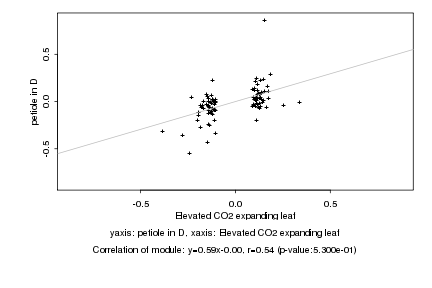
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Elevated CO2 expanding leaf |
Log2 signal ratio
petiole in D |
| 1 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.334 |
-0.009 |
| 2 |
257466_at |
AT1G62840
|
unknown |
0.251 |
-0.038 |
| 3 |
254571_at |
AT4G19370
|
unknown |
0.181 |
0.292 |
| 4 |
258201_at |
AT3G13910
|
unknown |
0.171 |
0.116 |
| 5 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.169 |
0.039 |
| 6 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.166 |
0.167 |
| 7 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.162 |
-0.061 |
| 8 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.152 |
0.868 |
| 9 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
0.148 |
0.114 |
| 10 |
259598_at |
AT1G27980
|
ATDPL1, DPL1, dihydrosphingosine phosphate lyase |
0.145 |
0.234 |
| 11 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.144 |
0.017 |
| 12 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.140 |
-0.000 |
| 13 |
250775_at |
AT5G05460
|
AtENGase85A, ENGase85A, Endo-beta-N-acetyglucosaminidase 85A |
0.136 |
0.105 |
| 14 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.134 |
0.027 |
| 15 |
257432_at |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.134 |
0.085 |
| 16 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
0.132 |
0.048 |
| 17 |
257928_at |
AT3G16890
|
PPR40, pentatricopeptide (PPR) domain protein 40 |
0.131 |
-0.045 |
| 18 |
252539_at |
AT3G45730
|
unknown |
0.127 |
0.223 |
| 19 |
265132_at |
AT1G23830
|
unknown |
0.126 |
-0.012 |
| 20 |
247570_at |
AT5G61250
|
AtGUS1, glucuronidase 1, GUS1, glucuronidase 1 |
0.125 |
0.077 |
| 21 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.123 |
0.075 |
| 22 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.123 |
0.039 |
| 23 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.122 |
-0.073 |
| 24 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
0.121 |
0.095 |
| 25 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.119 |
-0.063 |
| 26 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
0.116 |
0.032 |
| 27 |
252217_at |
AT3G50140
|
unknown |
0.115 |
0.124 |
| 28 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.114 |
0.184 |
| 29 |
264998_at |
AT1G67330
|
unknown |
0.113 |
-0.030 |
| 30 |
245775_at |
AT1G30270
|
ATCIPK23, CIPK23, CBL-interacting protein kinase 23, LKS1, LOW-K+-SENSITIVE 1, PKS17, SOS2-like protein kinase 17, SnRK3.23, SNF1-RELATED PROTEIN KINASE 3.23 |
0.113 |
0.081 |
| 31 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
0.110 |
-0.011 |
| 32 |
254752_at |
AT4G13160
|
unknown |
0.110 |
0.013 |
| 33 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.109 |
-0.200 |
| 34 |
256583_at |
AT3G28850
|
[Glutaredoxin family protein] |
0.109 |
0.047 |
| 35 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.108 |
0.244 |
| 36 |
248134_at |
AT5G54860
|
[Major facilitator superfamily protein] |
0.106 |
0.042 |
| 37 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
0.105 |
0.033 |
| 38 |
250881_at |
AT5G04080
|
unknown |
0.105 |
0.019 |
| 39 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.104 |
0.213 |
| 40 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
0.102 |
-0.043 |
| 41 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.100 |
0.120 |
| 42 |
250092_at |
AT5G17360
|
unknown |
0.099 |
0.028 |
| 43 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
0.096 |
0.139 |
| 44 |
256664_at |
AT3G12040
|
[DNA-3-methyladenine glycosylase (MAG)] |
0.096 |
0.125 |
| 45 |
262318_at |
AT1G27620
|
[HXXXD-type acyl-transferase family protein] |
0.096 |
0.026 |
| 46 |
259472_at |
AT1G18910
|
[zinc ion binding] |
0.094 |
0.049 |
| 47 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.094 |
-0.036 |
| 48 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
0.093 |
-0.030 |
| 49 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.089 |
-0.050 |
| 50 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.088 |
0.128 |
| 51 |
267459_at |
AT2G33850
|
unknown |
-0.386 |
-0.315 |
| 52 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.284 |
-0.353 |
| 53 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.243 |
-0.546 |
| 54 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.233 |
0.052 |
| 55 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.202 |
-0.200 |
| 56 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.199 |
-0.110 |
| 57 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.198 |
-0.144 |
| 58 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.187 |
-0.273 |
| 59 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.187 |
-0.041 |
| 60 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.184 |
-0.059 |
| 61 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.175 |
-0.036 |
| 62 |
251394_at |
AT3G60900
|
FLA10, FASCICLIN-like arabinogalactan-protein 10 |
-0.172 |
-0.067 |
| 63 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.172 |
0.007 |
| 64 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.157 |
0.083 |
| 65 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
-0.155 |
-0.021 |
| 66 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.152 |
-0.428 |
| 67 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
-0.149 |
0.056 |
| 68 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
-0.145 |
-0.045 |
| 69 |
256985_at |
AT3G13540
|
ATMYB5, myb domain protein 5, MYB5, myb domain protein 5 |
-0.145 |
-0.038 |
| 70 |
246063_at |
AT5G19340
|
unknown |
-0.145 |
0.032 |
| 71 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.144 |
-0.236 |
| 72 |
259714_at |
AT1G60980
|
ATGA20OX4, gibberellin 20-oxidase 4, GA20OX4, gibberellin 20-oxidase 4 |
-0.144 |
0.001 |
| 73 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.144 |
-0.118 |
| 74 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.143 |
-0.091 |
| 75 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.142 |
-0.053 |
| 76 |
246535_at |
AT5G15900
|
TBL19, TRICHOME BIREFRINGENCE-LIKE 19 |
-0.141 |
-0.063 |
| 77 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.137 |
-0.253 |
| 78 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.134 |
-0.010 |
| 79 |
249060_at |
AT5G44560
|
VPS2.2 |
-0.133 |
-0.112 |
| 80 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
-0.130 |
-0.121 |
| 81 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.130 |
-0.098 |
| 82 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.129 |
-0.020 |
| 83 |
262016_at |
AT1G35513
|
|
-0.128 |
0.065 |
| 84 |
257134_at |
AT3G12870
|
unknown |
-0.125 |
-0.073 |
| 85 |
265531_at |
AT2G06200
|
AtGRF6, growth-regulating factor 6, GRF6, growth-regulating factor 6 |
-0.121 |
0.026 |
| 86 |
262801_at |
AT1G21010
|
unknown |
-0.121 |
0.025 |
| 87 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.121 |
-0.128 |
| 88 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.121 |
0.233 |
| 89 |
252808_at |
AT3G42420
|
[pseudogene] |
-0.116 |
0.001 |
| 90 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
-0.116 |
0.012 |
| 91 |
262546_at |
AT1G31260
|
ZIP10, zinc transporter 10 precursor |
-0.114 |
-0.021 |
| 92 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.114 |
-0.089 |
| 93 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.114 |
-0.196 |
| 94 |
253064_at |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
-0.112 |
-0.081 |
| 95 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.110 |
-0.338 |
| 96 |
267552_at |
AT2G32770
|
ATPAP13, PURPLE ACID PHOSPHATASE 13, PAP13, purple acid phosphatase 13 |
-0.109 |
-0.005 |
| 97 |
261323_at |
AT1G44760
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.109 |
-0.091 |
| 98 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
-0.108 |
0.007 |
| 99 |
265555_at |
AT2G07500
|
[Mutator-like transposase family, has a 1.9e-70 P-value blast match to Q9SJR8 /172-333 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.107 |
0.029 |