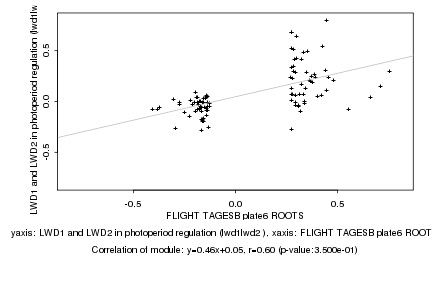
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
FLIGHT TAGESB plate6 ROOTS |
Log2 signal ratio
LWD1 and LWD2 in photoperiod regulation (lwd1lwd2 ) |
| 1 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
0.751 |
0.303 |
| 2 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.710 |
0.148 |
| 3 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.661 |
0.048 |
| 4 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
0.554 |
-0.077 |
| 5 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.477 |
0.209 |
| 6 |
245755_at |
AT1G35210
|
unknown |
0.453 |
0.246 |
| 7 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.445 |
0.111 |
| 8 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.443 |
0.807 |
| 9 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.441 |
0.310 |
| 10 |
260744_at |
AT1G15010
|
unknown |
0.425 |
0.549 |
| 11 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.421 |
0.066 |
| 12 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.399 |
0.058 |
| 13 |
260227_at |
AT1G74450
|
unknown |
0.390 |
0.246 |
| 14 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.384 |
0.272 |
| 15 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.373 |
0.192 |
| 16 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.370 |
0.256 |
| 17 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.366 |
0.198 |
| 18 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.362 |
0.213 |
| 19 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.350 |
0.497 |
| 20 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.348 |
0.286 |
| 21 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.341 |
0.132 |
| 22 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.338 |
-0.011 |
| 23 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.337 |
0.006 |
| 24 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.333 |
0.486 |
| 25 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.333 |
0.078 |
| 26 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
0.323 |
0.168 |
| 27 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.322 |
0.415 |
| 28 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.316 |
-0.089 |
| 29 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.312 |
0.077 |
| 30 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.309 |
-0.036 |
| 31 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.305 |
-0.047 |
| 32 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.297 |
0.648 |
| 33 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.296 |
0.429 |
| 34 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.292 |
0.061 |
| 35 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.292 |
0.290 |
| 36 |
263576_at |
AT2G17080
|
unknown |
0.291 |
-0.002 |
| 37 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.290 |
-0.036 |
| 38 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.285 |
0.421 |
| 39 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.284 |
0.354 |
| 40 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.282 |
0.518 |
| 41 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.280 |
0.300 |
| 42 |
249746_at |
AT5G24590
|
ANAC091, Arabidopsis NAC domain containing protein 91, TIP, TCV-interacting protein |
0.278 |
0.236 |
| 43 |
266037_at |
AT2G05940
|
RIPK, RPM1-induced protein kinase |
0.276 |
0.071 |
| 44 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.274 |
0.074 |
| 45 |
251259_at |
AT3G62260
|
[Protein phosphatase 2C family protein] |
0.273 |
0.136 |
| 46 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.273 |
0.531 |
| 47 |
265732_at |
AT2G01300
|
unknown |
0.272 |
0.341 |
| 48 |
247913_at |
AT5G57510
|
unknown |
0.272 |
0.015 |
| 49 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.271 |
0.682 |
| 50 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.271 |
-0.274 |
| 51 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.271 |
0.685 |
| 52 |
246018_at |
AT5G10695
|
unknown |
0.268 |
0.238 |
| 53 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.411 |
-0.078 |
| 54 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.383 |
-0.069 |
| 55 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.373 |
-0.050 |
| 56 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.306 |
0.022 |
| 57 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.296 |
-0.258 |
| 58 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.278 |
-0.026 |
| 59 |
247797_at |
AT5G58780
|
AtcPT5, AtHEPS, cPT5, cis -prenyltransferase 5, HEPS, heptaprenyl diphosphate synthase |
-0.275 |
-0.009 |
| 60 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.251 |
-0.107 |
| 61 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.227 |
-0.147 |
| 62 |
246872_at |
AT5G26080
|
[proline-rich family protein] |
-0.223 |
0.014 |
| 63 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.214 |
-0.021 |
| 64 |
256097_at |
AT1G13670
|
unknown |
-0.203 |
-0.005 |
| 65 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
-0.200 |
0.097 |
| 66 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
-0.198 |
-0.095 |
| 67 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.192 |
0.047 |
| 68 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.190 |
0.049 |
| 69 |
252882_at |
AT4G39675
|
unknown |
-0.189 |
-0.075 |
| 70 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
-0.189 |
-0.001 |
| 71 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.185 |
-0.022 |
| 72 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.181 |
-0.061 |
| 73 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
-0.178 |
-0.078 |
| 74 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.177 |
0.005 |
| 75 |
251335_at |
AT3G61400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.176 |
0.007 |
| 76 |
250640_at |
AT5G07150
|
[Leucine-rich repeat protein kinase family protein] |
-0.175 |
-0.046 |
| 77 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.171 |
-0.280 |
| 78 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.171 |
-0.169 |
| 79 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-0.169 |
-0.057 |
| 80 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.169 |
-0.098 |
| 81 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-0.166 |
-0.006 |
| 82 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
-0.165 |
-0.175 |
| 83 |
252010_at |
AT3G52740
|
unknown |
-0.165 |
-0.181 |
| 84 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.162 |
-0.194 |
| 85 |
261247_at |
AT1G20070
|
unknown |
-0.161 |
-0.163 |
| 86 |
249138_at |
AT5G43070
|
WPP1, WPP domain protein 1 |
-0.161 |
-0.005 |
| 87 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.160 |
0.030 |
| 88 |
251727_at |
AT3G56290
|
unknown |
-0.157 |
-0.187 |
| 89 |
251293_at |
AT3G61930
|
unknown |
-0.152 |
-0.050 |
| 90 |
258770_at |
AT3G10820
|
[Transcription elongation factor (TFIIS) family protein] |
-0.147 |
0.032 |
| 91 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
-0.146 |
-0.067 |
| 92 |
262590_at |
AT1G15100
|
RHA2A, RING-H2 finger A2A |
-0.144 |
-0.136 |
| 93 |
247807_at |
AT5G58360
|
ATOFP3, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 3, OFP3, ovate family protein 3 |
-0.143 |
0.065 |
| 94 |
253513_at |
AT4G31760
|
[Peroxidase superfamily protein] |
-0.143 |
0.044 |
| 95 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.143 |
-0.085 |
| 96 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.142 |
0.051 |
| 97 |
262322_at |
AT1G27590
|
unknown |
-0.141 |
-0.043 |
| 98 |
260300_at |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
-0.140 |
0.052 |
| 99 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
-0.138 |
-0.083 |
| 100 |
255745_at |
AT1G32030
|
[FUNCTIONS IN: DNA binding] |
-0.138 |
-0.002 |
| 101 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.136 |
-0.252 |
| 102 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.131 |
-0.042 |
| 103 |
247473_at |
AT5G62260
|
AHL6, AT-hook motif nuclear localized protein 6 |
-0.130 |
-0.014 |