|
probeID |
AGICode |
Annotation |
Log2 signal ratio
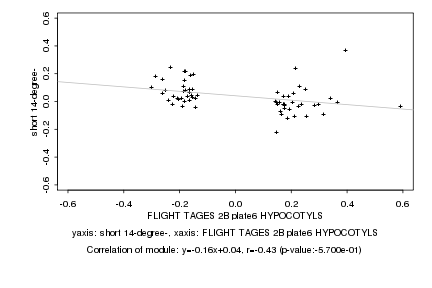
FLIGHT TAGES 2B plate6 HYPOCOTYLS |
Log2 signal ratio
short 14-degree- |
| 1 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.590 |
-0.035 |
| 2 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.394 |
0.370 |
| 3 |
255754_at |
AT1G43040
|
SAUR58, SMALL AUXIN UPREGULATED RNA 58 |
0.363 |
-0.005 |
| 4 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
0.340 |
0.024 |
| 5 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.315 |
-0.089 |
| 6 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.297 |
-0.017 |
| 7 |
249896_at |
AT5G22530
|
unknown |
0.282 |
-0.022 |
| 8 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.253 |
-0.101 |
| 9 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.248 |
0.092 |
| 10 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.236 |
-0.018 |
| 11 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.226 |
0.114 |
| 12 |
249393_at |
AT5G40170
|
AtRLP54, receptor like protein 54, RLP54, receptor like protein 54 |
0.225 |
-0.030 |
| 13 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.213 |
0.244 |
| 14 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.211 |
-0.102 |
| 15 |
253067_at |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
0.207 |
0.063 |
| 16 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
0.203 |
-0.001 |
| 17 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.191 |
-0.051 |
| 18 |
251723_at |
AT3G56230
|
[BTB/POZ domain-containing protein] |
0.187 |
0.037 |
| 19 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.185 |
-0.122 |
| 20 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.175 |
-0.023 |
| 21 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.175 |
-0.017 |
| 22 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.174 |
-0.050 |
| 23 |
261748_at |
AT1G76070
|
unknown |
0.172 |
-0.017 |
| 24 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
0.171 |
0.042 |
| 25 |
266111_at |
AT2G02061
|
[Nucleotide-diphospho-sugar transferase family protein] |
0.170 |
-0.019 |
| 26 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.165 |
-0.090 |
| 27 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
0.160 |
-0.068 |
| 28 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
0.155 |
-0.006 |
| 29 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.150 |
0.071 |
| 30 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.149 |
-0.019 |
| 31 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.146 |
-0.221 |
| 32 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.145 |
-0.002 |
| 33 |
255766_at |
AT1G16750
|
unknown |
0.143 |
0.002 |
| 34 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.303 |
0.108 |
| 35 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.290 |
0.181 |
| 36 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.263 |
0.161 |
| 37 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.263 |
0.063 |
| 38 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
-0.254 |
0.086 |
| 39 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.240 |
0.012 |
| 40 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.233 |
0.245 |
| 41 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.229 |
-0.018 |
| 42 |
248862_at |
AT5G46730
|
[glycine-rich protein] |
-0.224 |
0.038 |
| 43 |
254352_at |
AT4G22320
|
unknown |
-0.208 |
0.025 |
| 44 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.207 |
0.018 |
| 45 |
262033_at |
AT1G37140
|
MCT1, MEI2 C-terminal RRM only like 1 |
-0.195 |
0.027 |
| 46 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.192 |
-0.031 |
| 47 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.189 |
0.110 |
| 48 |
256799_at |
AT3G18560
|
unknown |
-0.188 |
0.079 |
| 49 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.186 |
0.222 |
| 50 |
265482_at |
AT2G15780
|
[Cupredoxin superfamily protein] |
-0.185 |
0.004 |
| 51 |
252010_at |
AT3G52740
|
unknown |
-0.183 |
0.154 |
| 52 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.180 |
0.085 |
| 53 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.180 |
0.218 |
| 54 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.175 |
0.039 |
| 55 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
-0.168 |
0.069 |
| 56 |
251727_at |
AT3G56290
|
unknown |
-0.168 |
0.087 |
| 57 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
-0.165 |
0.012 |
| 58 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.164 |
0.190 |
| 59 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.160 |
0.044 |
| 60 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
-0.156 |
0.090 |
| 61 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.155 |
0.029 |
| 62 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.151 |
0.195 |
| 63 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.144 |
-0.037 |
| 64 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
-0.144 |
0.027 |
| 65 |
267226_at |
AT2G44010
|
unknown |
-0.138 |
0.046 |