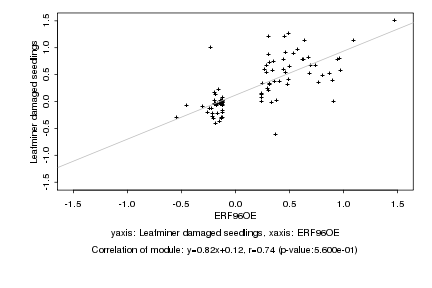
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ERF96OE |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
1.465 |
1.524 |
| 2 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.087 |
1.150 |
| 3 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.967 |
0.580 |
| 4 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.956 |
0.813 |
| 5 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.941 |
0.792 |
| 6 |
249154_at |
AT5G43410
|
[Integrase-type DNA-binding superfamily protein] |
0.902 |
0.005 |
| 7 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.897 |
0.393 |
| 8 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.868 |
0.524 |
| 9 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.797 |
0.497 |
| 10 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
0.760 |
0.364 |
| 11 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.736 |
0.687 |
| 12 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.694 |
0.685 |
| 13 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.682 |
0.538 |
| 14 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.671 |
0.836 |
| 15 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.630 |
1.152 |
| 16 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.621 |
0.785 |
| 17 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.619 |
0.789 |
| 18 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.572 |
0.971 |
| 19 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.537 |
0.902 |
| 20 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.492 |
0.654 |
| 21 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.487 |
1.277 |
| 22 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.483 |
0.425 |
| 23 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
0.475 |
0.330 |
| 24 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.457 |
0.916 |
| 25 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.456 |
0.557 |
| 26 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.451 |
1.221 |
| 27 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.444 |
0.795 |
| 28 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.440 |
0.609 |
| 29 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.404 |
0.382 |
| 30 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.371 |
0.024 |
| 31 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.368 |
-0.611 |
| 32 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.356 |
0.387 |
| 33 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.352 |
0.747 |
| 34 |
263032_at |
AT1G23850
|
unknown |
0.338 |
0.580 |
| 35 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
0.327 |
-0.010 |
| 36 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.309 |
0.339 |
| 37 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.309 |
0.318 |
| 38 |
245545_at |
AT4G15280
|
UGT71B5, UDP-glucosyl transferase 71B5 |
0.308 |
0.740 |
| 39 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.305 |
0.877 |
| 40 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.304 |
1.212 |
| 41 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.302 |
0.207 |
| 42 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.295 |
0.246 |
| 43 |
263904_at |
AT2G36380
|
ABCG34, ATP-binding cassette G34, ATPDR6, PLEIOTROPIC DRUG RESISTANCE 6, PDR6, pleiotropic drug resistance 6 |
0.286 |
0.550 |
| 44 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.283 |
0.672 |
| 45 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.263 |
0.605 |
| 46 |
267610_at |
AT2G26650
|
AKT1, K+ transporter 1, ATAKT1, KT1, K+ transporter 1 |
0.245 |
0.339 |
| 47 |
252054_at |
AT3G52540
|
ATOFP18, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 18, OFP18, ovate family protein 18 |
0.240 |
0.018 |
| 48 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.236 |
0.147 |
| 49 |
257321_at |
ATMG01130
|
ORF106F |
0.236 |
0.079 |
| 50 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.234 |
0.156 |
| 51 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.547 |
-0.287 |
| 52 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.455 |
-0.066 |
| 53 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-0.310 |
-0.076 |
| 54 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.266 |
-0.203 |
| 55 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
-0.245 |
-0.125 |
| 56 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.234 |
1.005 |
| 57 |
257761_at |
AT3G23090
|
WDL3 |
-0.226 |
-0.128 |
| 58 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.220 |
-0.277 |
| 59 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.215 |
-0.210 |
| 60 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.210 |
-0.311 |
| 61 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.202 |
0.034 |
| 62 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.200 |
0.174 |
| 63 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.199 |
-0.033 |
| 64 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
-0.194 |
-0.055 |
| 65 |
256066_at |
AT1G06980
|
unknown |
-0.187 |
-0.405 |
| 66 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.186 |
-0.035 |
| 67 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.185 |
0.136 |
| 68 |
248586_at |
AT5G49610
|
[F-box family protein] |
-0.177 |
-0.056 |
| 69 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.172 |
-0.214 |
| 70 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.171 |
-0.029 |
| 71 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.160 |
0.238 |
| 72 |
248616_at |
AT5G49580
|
[Chaperone DnaJ-domain superfamily protein] |
-0.152 |
-0.036 |
| 73 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.152 |
-0.355 |
| 74 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.152 |
-0.026 |
| 75 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.147 |
-0.011 |
| 76 |
250431_at |
AT5G10440
|
CYCD4;2, cyclin d4;2 |
-0.147 |
0.033 |
| 77 |
267453_at |
AT2G33880
|
HB-3, homeobox-3, STIP, STIMPY, WOX9, WUSCHEL-RELATED HOMEOBOX 9, WOX9A, WUSCHEL related homeobox 9A |
-0.139 |
-0.045 |
| 78 |
248993_at |
AT5G45240
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.138 |
0.017 |
| 79 |
252910_at |
AT4G39590
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.138 |
-0.032 |
| 80 |
254155_at |
AT4G24480
|
[Protein kinase superfamily protein] |
-0.137 |
-0.036 |
| 81 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.137 |
-0.032 |
| 82 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
-0.135 |
-0.007 |
| 83 |
266088_at |
AT2G37780
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.134 |
0.032 |
| 84 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
-0.133 |
-0.309 |
| 85 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.133 |
-0.281 |
| 86 |
261434_at |
AT1G07650
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.130 |
-0.018 |
| 87 |
265903_at |
AT2G25600
|
AKT6, SPIK, Shaker pollen inward K+ channel |
-0.130 |
-0.027 |
| 88 |
264480_at |
AT1G77300
|
ASHH2, ASH1 HOMOLOG 2, CCR1, CAROTENOID CHLOROPLAST REGULATORY1, EFS, EARLY FLOWERING IN SHORT DAYS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 |
-0.130 |
-0.039 |
| 89 |
254753_at |
AT4G13190
|
[Protein kinase superfamily protein] |
-0.129 |
0.023 |
| 90 |
251058_at |
AT5G01790
|
unknown |
-0.129 |
-0.294 |
| 91 |
262764_at |
AT1G13140
|
CYP86C3, cytochrome P450, family 86, subfamily C, polypeptide 3 |
-0.129 |
0.078 |
| 92 |
253839_at |
AT4G27890
|
[HSP20-like chaperones superfamily protein] |
-0.128 |
-0.015 |
| 93 |
265833_at |
AT2G14420
|
[Mutator-like transposase family, has a 8.4e-10 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.127 |
0.001 |
| 94 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.127 |
-0.160 |
| 95 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.126 |
-0.190 |
| 96 |
249921_at |
AT5G19270
|
unknown |
-0.126 |
0.020 |
| 97 |
246911_at |
AT5G25810
|
tny, TINY |
-0.125 |
-0.000 |
| 98 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
-0.125 |
-0.042 |
| 99 |
248051_at |
AT5G56110
|
AtMYB103, myb domain protein 103, ATMYB80, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 80, MS188, MALE STERILE 188, MYB103, myb domain protein 103, MYB80 |
-0.125 |
-0.058 |