|
probeID |
AGICode |
Annotation |
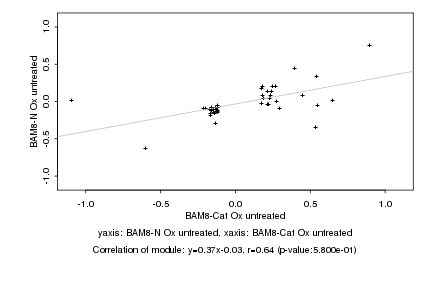
Log2 signal ratio
BAM8-Cat Ox untreated |
Log2 signal ratio
BAM8-N Ox untreated |
| 1 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.891 |
0.757 |
| 2 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
0.647 |
0.027 |
| 3 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
0.546 |
-0.048 |
| 4 |
247882_at |
AT5G57785
|
unknown |
0.536 |
0.345 |
| 5 |
248997_at |
AT5G45300
|
BAM8, BETA-AMYLASE 8, BMY2, beta-amylase 2 |
0.530 |
-0.336 |
| 6 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.443 |
0.085 |
| 7 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.393 |
0.443 |
| 8 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.293 |
-0.091 |
| 9 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.270 |
0.012 |
| 10 |
255942_at |
AT1G22360
|
AtUGT85A2, UDP-glucosyl transferase 85A2, UGT85A2, UDP-glucosyl transferase 85A2 |
0.264 |
0.209 |
| 11 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.244 |
0.203 |
| 12 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.236 |
0.144 |
| 13 |
246235_at |
AT4G36830
|
HOS3-1 |
0.228 |
0.081 |
| 14 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
0.222 |
0.042 |
| 15 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.220 |
-0.033 |
| 16 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
0.212 |
0.145 |
| 17 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.210 |
-0.028 |
| 18 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.186 |
0.051 |
| 19 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.178 |
0.083 |
| 20 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.175 |
0.212 |
| 21 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
0.174 |
-0.019 |
| 22 |
247105_at |
AT5G66630
|
DAR5, DA1-related protein 5 |
0.174 |
0.182 |
| 23 |
252606_at |
AT3G45010
|
scpl48, serine carboxypeptidase-like 48 |
-1.100 |
0.021 |
| 24 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.604 |
-0.619 |
| 25 |
252446_at |
AT3G47010
|
[Glycosyl hydrolase family protein] |
-0.218 |
-0.090 |
| 26 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.202 |
-0.082 |
| 27 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
-0.178 |
-0.104 |
| 28 |
260021_at |
AT1G30010
|
nMAT1, nuclear maturase 1 |
-0.168 |
-0.149 |
| 29 |
251350_at |
AT3G61040
|
CYP76C7, cytochrome P450, family 76, subfamily C, polypeptide 7 |
-0.167 |
-0.179 |
| 30 |
249261_at |
AT5G41710
|
[Mutator-like transposase family, has a 6.2e-18 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.166 |
-0.113 |
| 31 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
-0.164 |
-0.079 |
| 32 |
257524_at |
AT3G01330
|
DEL3, DP-E2F-like protein 3, E2FF, E2L2, E2F-LIKE 2 |
-0.153 |
-0.106 |
| 33 |
250622_at |
AT5G07310
|
[Integrase-type DNA-binding superfamily protein] |
-0.147 |
-0.136 |
| 34 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
-0.142 |
-0.158 |
| 35 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.139 |
-0.293 |
| 36 |
256896_at |
AT3G24630
|
TRM34, TON1 Recruiting Motif 34 |
-0.138 |
-0.070 |
| 37 |
256871_at |
AT3G26480
|
[Transducin family protein / WD-40 repeat family protein] |
-0.129 |
-0.129 |
| 38 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.127 |
-0.101 |
| 39 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.126 |
-0.070 |
| 40 |
260688_at |
AT1G17665
|
unknown |
-0.126 |
-0.041 |
| 41 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.125 |
-0.113 |
| 42 |
253472_at |
AT4G32230
|
unknown |
-0.125 |
-0.142 |
| 43 |
255900_at |
AT1G17830
|
unknown |
-0.124 |
-0.129 |