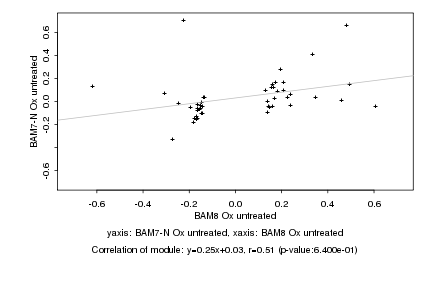
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
BAM8 Ox untreated |
Log2 signal ratio
BAM7-N Ox untreated |
| 1 |
248997_at |
AT5G45300
|
BAM8, BETA-AMYLASE 8, BMY2, beta-amylase 2 |
0.604 |
-0.041 |
| 2 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
0.492 |
0.151 |
| 3 |
247882_at |
AT5G57785
|
unknown |
0.480 |
0.666 |
| 4 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
0.458 |
0.011 |
| 5 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
0.343 |
0.039 |
| 6 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.332 |
0.413 |
| 7 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
0.236 |
0.067 |
| 8 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.235 |
-0.029 |
| 9 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.222 |
0.039 |
| 10 |
252590_at |
AT3G45670
|
[Protein kinase superfamily protein] |
0.208 |
0.104 |
| 11 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.205 |
0.167 |
| 12 |
256789_at |
AT3G13672
|
SINA2 |
0.193 |
0.286 |
| 13 |
249952_at |
AT5G18950
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.178 |
0.090 |
| 14 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.171 |
0.166 |
| 15 |
266487_at |
AT2G47660
|
unknown |
0.168 |
0.034 |
| 16 |
258137_at |
AT3G24515
|
UBC37, ubiquitin-conjugating enzyme 37 |
0.163 |
0.126 |
| 17 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.159 |
-0.038 |
| 18 |
250585_at |
AT5G07620
|
[Protein kinase superfamily protein] |
0.158 |
0.155 |
| 19 |
254807_at |
AT4G12700
|
unknown |
0.155 |
0.127 |
| 20 |
259105_at |
AT3G05500
|
[Rubber elongation factor protein (REF)] |
0.144 |
-0.049 |
| 21 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.143 |
-0.036 |
| 22 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
0.137 |
-0.092 |
| 23 |
256082_at |
AT1G20720
|
[RAD3-like DNA-binding helicase protein] |
0.137 |
0.005 |
| 24 |
262150_at |
AT1G52520
|
FRS6, FAR1-related sequence 6 |
0.129 |
0.100 |
| 25 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.620 |
0.138 |
| 26 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.309 |
0.075 |
| 27 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.273 |
-0.330 |
| 28 |
261292_at |
AT1G36940
|
unknown |
-0.249 |
-0.010 |
| 29 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.226 |
0.713 |
| 30 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.195 |
-0.047 |
| 31 |
250020_at |
AT5G18180
|
[H/ACA ribonucleoprotein complex, subunit Gar1/Naf1 protein] |
-0.182 |
-0.179 |
| 32 |
256871_at |
AT3G26480
|
[Transducin family protein / WD-40 repeat family protein] |
-0.179 |
-0.146 |
| 33 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.172 |
-0.148 |
| 34 |
267409_at |
AT2G34910
|
unknown |
-0.170 |
-0.128 |
| 35 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.168 |
-0.078 |
| 36 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.165 |
-0.142 |
| 37 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
-0.165 |
-0.020 |
| 38 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.165 |
-0.056 |
| 39 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.159 |
-0.065 |
| 40 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.155 |
-0.031 |
| 41 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.152 |
-0.054 |
| 42 |
255533_at |
AT4G02180
|
[DC1 domain-containing protein] |
-0.150 |
-0.103 |
| 43 |
259552_at |
AT1G21320
|
[nucleotide binding] |
-0.149 |
-0.007 |
| 44 |
252569_at |
AT3G45420
|
LecRK-I.4, L-type lectin receptor kinase I.4 |
-0.146 |
-0.040 |
| 45 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.145 |
-0.099 |
| 46 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
-0.140 |
0.040 |
| 47 |
266971_at |
AT2G39580
|
unknown |
-0.136 |
0.035 |