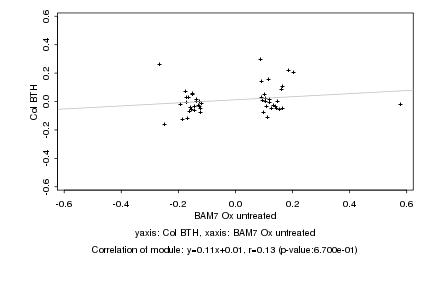
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
BAM7 Ox untreated |
Log2 signal ratio
Col BTH |
| 1 |
266914_at |
AT2G45880
|
BAM7, beta-amylase 7, BMY4, BETA-AMYLASE 4 |
0.577 |
-0.017 |
| 2 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.201 |
0.206 |
| 3 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.185 |
0.222 |
| 4 |
258157_at |
AT3G18100
|
AtMYB4R1, myb domain protein 4R1, MYB4R1, myb domain protein 4r1 |
0.163 |
0.107 |
| 5 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
0.162 |
-0.046 |
| 6 |
245076_at |
AT2G23170
|
GH3.3 |
0.160 |
0.086 |
| 7 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.153 |
-0.055 |
| 8 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
0.147 |
0.003 |
| 9 |
247504_at |
AT5G61990
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.142 |
-0.046 |
| 10 |
257725_at |
AT3G18524
|
ATMSH2, MSH2, MUTS homolog 2 |
0.140 |
-0.028 |
| 11 |
256404_at |
AT3G06250
|
FRS7, FAR1-related sequence 7 |
0.133 |
-0.024 |
| 12 |
247206_at |
AT5G64950
|
[Mitochondrial transcription termination factor family protein] |
0.123 |
-0.049 |
| 13 |
254054_at |
AT4G25320
|
AHL3, AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 3 |
0.116 |
0.015 |
| 14 |
259973_at |
AT1G76630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.116 |
-0.004 |
| 15 |
252879_at |
AT4G39390
|
ATNST-KT1, A. THALIANA NUCLEOTIDE SUGAR TRANSPORTER-KT 1, NST-K1, nucleotide sugar transporter-KT 1 |
0.113 |
0.155 |
| 16 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.112 |
-0.107 |
| 17 |
262415_at |
AT1G49400
|
emb1129, embryo defective 1129 |
0.106 |
-0.030 |
| 18 |
254812_at |
AT4G12250
|
GAE5, UDP-D-glucuronate 4-epimerase 5 |
0.103 |
0.023 |
| 19 |
267613_at |
AT2G26700
|
PID2, PINOID2 |
0.102 |
0.006 |
| 20 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.100 |
0.055 |
| 21 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
0.095 |
-0.074 |
| 22 |
267004_at |
AT2G34260
|
WDR55, human WDR55 (WD40 repeat) homolog |
0.094 |
0.009 |
| 23 |
250543_at |
AT5G08190
|
NF-YB12, nuclear factor Y, subunit B12 |
0.090 |
0.032 |
| 24 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.088 |
0.144 |
| 25 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.087 |
0.296 |
| 26 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.269 |
0.267 |
| 27 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.249 |
-0.159 |
| 28 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.194 |
-0.020 |
| 29 |
247903_at |
AT5G57340
|
unknown |
-0.186 |
-0.120 |
| 30 |
265009_at |
AT1G24650
|
[Leucine-rich repeat protein kinase family protein] |
-0.175 |
0.071 |
| 31 |
250699_at |
AT5G06820
|
SRF2, STRUBBELIG-receptor family 2 |
-0.173 |
0.035 |
| 32 |
259377_at |
AT3G16380
|
PAB6, poly(A) binding protein 6 |
-0.172 |
-0.003 |
| 33 |
256408_at |
AT1G66610
|
[TRAF-like superfamily protein] |
-0.169 |
-0.117 |
| 34 |
247253_at |
AT5G64790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.167 |
0.034 |
| 35 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.163 |
-0.065 |
| 36 |
263977_at |
AT2G42660
|
[Homeodomain-like superfamily protein] |
-0.158 |
-0.039 |
| 37 |
250937_at |
AT5G03230
|
unknown |
-0.155 |
-0.056 |
| 38 |
259946_at |
AT1G71450
|
[Integrase-type DNA-binding superfamily protein] |
-0.151 |
0.059 |
| 39 |
262049_at |
AT1G80180
|
unknown |
-0.151 |
0.050 |
| 40 |
257837_at |
AT3G25200
|
unknown |
-0.146 |
-0.060 |
| 41 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.145 |
-0.032 |
| 42 |
250271_at |
AT5G12990
|
CLE40, CLAVATA3/ESR-RELATED 40 |
-0.138 |
0.005 |
| 43 |
251382_at |
AT3G60730
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.138 |
0.018 |
| 44 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-0.131 |
-0.021 |
| 45 |
265645_at |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
-0.127 |
-0.032 |
| 46 |
254989_at |
AT4G10690
|
[copia-like retrotransposon family, has a 1.5e-256 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.126 |
0.003 |
| 47 |
257683_at |
AT3G13280
|
[Putative endonuclease or glycosyl hydrolase] |
-0.125 |
-0.043 |
| 48 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.124 |
-0.071 |
| 49 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
-0.122 |
-0.010 |