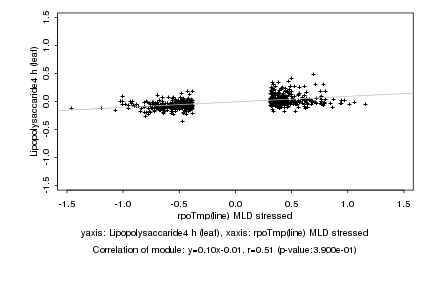
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rpoTmp(line) MLD stressed |
Log2 signal ratio
Lipopolysaccaride4 h (leaf) |
| 1 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
1.156 |
-0.039 |
| 2 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
1.059 |
-0.002 |
| 3 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.013 |
-0.044 |
| 4 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.967 |
0.035 |
| 5 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.944 |
0.026 |
| 6 |
265954_at |
AT2G37260
|
ATWRKY44, DSL1, DR. STRANGELOVE 1, TTG2, TRANSPARENT TESTA GLABRA 2, WRKY44 |
0.936 |
-0.020 |
| 7 |
249454_at |
AT5G39520
|
unknown |
0.929 |
-0.028 |
| 8 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.866 |
0.042 |
| 9 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.857 |
-0.099 |
| 10 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.845 |
-0.035 |
| 11 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.797 |
0.038 |
| 12 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.793 |
0.180 |
| 13 |
251012_at |
AT5G02580
|
unknown |
0.792 |
-0.000 |
| 14 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.787 |
-0.032 |
| 15 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.780 |
0.310 |
| 16 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.779 |
-0.060 |
| 17 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.765 |
-0.041 |
| 18 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.763 |
0.015 |
| 19 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.759 |
0.037 |
| 20 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.758 |
0.106 |
| 21 |
256924_at |
AT3G29590
|
AT5MAT |
0.758 |
0.015 |
| 22 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.757 |
0.187 |
| 23 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.751 |
0.072 |
| 24 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.749 |
-0.024 |
| 25 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.720 |
0.057 |
| 26 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.714 |
-0.028 |
| 27 |
258203_at |
AT3G13950
|
unknown |
0.704 |
0.317 |
| 28 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.704 |
-0.005 |
| 29 |
256789_at |
AT3G13672
|
SINA2 |
0.702 |
-0.000 |
| 30 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.691 |
0.493 |
| 31 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.685 |
0.006 |
| 32 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.683 |
0.009 |
| 33 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.683 |
-0.014 |
| 34 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.676 |
0.022 |
| 35 |
244931_at |
ATMG00630
|
ORF110B |
0.672 |
-0.036 |
| 36 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.666 |
0.025 |
| 37 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.658 |
0.034 |
| 38 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.652 |
0.040 |
| 39 |
257323_at |
ATMG01200
|
ORF294 |
0.652 |
0.027 |
| 40 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.640 |
-0.041 |
| 41 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
0.639 |
-0.036 |
| 42 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.637 |
-0.089 |
| 43 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.635 |
-0.105 |
| 44 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.635 |
-0.030 |
| 45 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.627 |
0.068 |
| 46 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.627 |
0.165 |
| 47 |
267646_at |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
0.626 |
0.044 |
| 48 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.624 |
-0.002 |
| 49 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.623 |
0.034 |
| 50 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.617 |
-0.108 |
| 51 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.614 |
-0.046 |
| 52 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.611 |
-0.081 |
| 53 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.611 |
0.108 |
| 54 |
254853_at |
AT4G12080
|
AHL1, AT-hook motif nuclear-localized protein 1, ATAHL1 |
0.606 |
0.035 |
| 55 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.606 |
0.282 |
| 56 |
263881_at |
AT2G21820
|
unknown |
0.593 |
-0.029 |
| 57 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.592 |
0.078 |
| 58 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.590 |
0.054 |
| 59 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
0.573 |
0.012 |
| 60 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.568 |
0.253 |
| 61 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.562 |
0.157 |
| 62 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.562 |
-0.016 |
| 63 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.559 |
-0.095 |
| 64 |
254571_at |
AT4G19370
|
unknown |
0.559 |
0.120 |
| 65 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.555 |
0.141 |
| 66 |
254178_at |
AT4G23880
|
unknown |
0.553 |
0.019 |
| 67 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.551 |
-0.047 |
| 68 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.546 |
-0.006 |
| 69 |
261134_at |
AT1G19630
|
CYP722A1, cytochrome P450, family 722, subfamily A, polypeptide 1 |
0.539 |
-0.042 |
| 70 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.533 |
-0.162 |
| 71 |
252400_at |
AT3G48020
|
unknown |
0.531 |
-0.045 |
| 72 |
262719_at |
AT1G43590
|
unknown |
0.526 |
0.006 |
| 73 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.525 |
0.000 |
| 74 |
257322_at |
ATMG01180
|
ORF111B |
0.523 |
0.015 |
| 75 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.518 |
0.271 |
| 76 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.517 |
0.105 |
| 77 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.516 |
0.049 |
| 78 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.511 |
-0.052 |
| 79 |
264590_at |
AT2G17710
|
unknown |
0.506 |
0.064 |
| 80 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.504 |
0.005 |
| 81 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.497 |
0.017 |
| 82 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.496 |
0.412 |
| 83 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.495 |
0.189 |
| 84 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
0.493 |
0.141 |
| 85 |
265059_at |
AT1G52080
|
AR791 |
0.493 |
0.093 |
| 86 |
264287_at |
AT1G61930
|
unknown |
0.488 |
-0.022 |
| 87 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.486 |
-0.014 |
| 88 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.485 |
0.284 |
| 89 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.483 |
0.283 |
| 90 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.483 |
-0.045 |
| 91 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.481 |
0.220 |
| 92 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.474 |
0.144 |
| 93 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.469 |
0.360 |
| 94 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
0.469 |
-0.078 |
| 95 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
0.468 |
0.170 |
| 96 |
260522_x_at |
AT2G41730
|
unknown |
0.467 |
0.115 |
| 97 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.466 |
-0.013 |
| 98 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.465 |
-0.012 |
| 99 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.464 |
0.035 |
| 100 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.463 |
0.171 |
| 101 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.461 |
0.187 |
| 102 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
0.460 |
-0.022 |
| 103 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.459 |
-0.033 |
| 104 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.458 |
-0.090 |
| 105 |
265913_at |
AT2G25625
|
unknown |
0.458 |
-0.046 |
| 106 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.457 |
0.029 |
| 107 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.457 |
-0.050 |
| 108 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.456 |
-0.034 |
| 109 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.456 |
0.171 |
| 110 |
245662_at |
AT1G28190
|
unknown |
0.452 |
0.095 |
| 111 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.451 |
0.001 |
| 112 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.450 |
0.023 |
| 113 |
247751_at |
AT5G59050
|
unknown |
0.450 |
0.055 |
| 114 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.448 |
-0.023 |
| 115 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.448 |
-0.009 |
| 116 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.447 |
-0.042 |
| 117 |
247800_at |
AT5G58570
|
unknown |
0.443 |
0.099 |
| 118 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.442 |
0.079 |
| 119 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.441 |
0.204 |
| 120 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.441 |
0.197 |
| 121 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.440 |
0.010 |
| 122 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.440 |
0.028 |
| 123 |
266488_at |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
0.439 |
-0.052 |
| 124 |
247212_at |
AT5G65040
|
unknown |
0.439 |
-0.003 |
| 125 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.437 |
0.082 |
| 126 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.437 |
0.234 |
| 127 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.436 |
0.095 |
| 128 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.436 |
-0.105 |
| 129 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
0.435 |
-0.004 |
| 130 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.435 |
-0.015 |
| 131 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.434 |
0.020 |
| 132 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.433 |
-0.164 |
| 133 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.433 |
0.013 |
| 134 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
0.432 |
0.001 |
| 135 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.432 |
0.016 |
| 136 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.431 |
0.089 |
| 137 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.428 |
-0.060 |
| 138 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.425 |
0.109 |
| 139 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.425 |
0.051 |
| 140 |
259424_at |
AT1G13830
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.425 |
-0.025 |
| 141 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.424 |
0.148 |
| 142 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.417 |
0.103 |
| 143 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.417 |
0.121 |
| 144 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.417 |
-0.010 |
| 145 |
265356_at |
AT2G16595
|
[Translocon-associated protein (TRAP), alpha subunit] |
0.414 |
0.262 |
| 146 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.413 |
-0.052 |
| 147 |
255879_at |
AT1G67000
|
[Protein kinase superfamily protein] |
0.411 |
0.021 |
| 148 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.411 |
0.018 |
| 149 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.409 |
0.105 |
| 150 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.408 |
0.227 |
| 151 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.408 |
-0.103 |
| 152 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.407 |
-0.058 |
| 153 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.407 |
0.043 |
| 154 |
262452_at |
AT1G11210
|
unknown |
0.406 |
0.051 |
| 155 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.406 |
0.063 |
| 156 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.405 |
-0.091 |
| 157 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.404 |
0.234 |
| 158 |
253137_at |
AT4G35500
|
[Protein kinase superfamily protein] |
0.401 |
0.033 |
| 159 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.401 |
0.058 |
| 160 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.401 |
0.056 |
| 161 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.401 |
-0.095 |
| 162 |
246125_at |
AT5G19875
|
unknown |
0.398 |
0.062 |
| 163 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.398 |
-0.030 |
| 164 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.396 |
0.010 |
| 165 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.394 |
-0.076 |
| 166 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.393 |
0.000 |
| 167 |
253827_at |
AT4G28085
|
unknown |
0.392 |
-0.110 |
| 168 |
267413_at |
AT2G34960
|
CAT5, cationic amino acid transporter 5 |
0.391 |
-0.039 |
| 169 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.391 |
0.017 |
| 170 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.390 |
0.115 |
| 171 |
255912_at |
AT1G66960
|
LUP5 |
0.388 |
0.011 |
| 172 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.388 |
-0.011 |
| 173 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.388 |
0.147 |
| 174 |
258362_at |
AT3G14280
|
unknown |
0.386 |
0.090 |
| 175 |
264851_at |
AT2G17290
|
ATCDPK3, ARABIDOPSIS THALIANA CALMODULIN-DOMAIN PROTEIN KINASE 3, ATCPK6, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 6, CPK6, calcium dependent protein kinase 6 |
0.386 |
0.212 |
| 176 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.385 |
-0.082 |
| 177 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.385 |
-0.043 |
| 178 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.385 |
-0.026 |
| 179 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.384 |
-0.050 |
| 180 |
256188_at |
AT1G30160
|
unknown |
0.384 |
-0.023 |
| 181 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.381 |
0.046 |
| 182 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
0.381 |
0.057 |
| 183 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.381 |
0.349 |
| 184 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.381 |
0.167 |
| 185 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.380 |
0.031 |
| 186 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.380 |
0.054 |
| 187 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.380 |
-0.049 |
| 188 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.379 |
0.079 |
| 189 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
0.378 |
-0.016 |
| 190 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.378 |
-0.004 |
| 191 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.376 |
-0.034 |
| 192 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
0.376 |
-0.005 |
| 193 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.376 |
0.139 |
| 194 |
249134_at |
AT5G43150
|
unknown |
0.376 |
-0.095 |
| 195 |
251984_at |
AT3G53260
|
ATPAL2, PAL2, phenylalanine ammonia-lyase 2 |
0.375 |
0.053 |
| 196 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.374 |
0.083 |
| 197 |
265892_at |
AT2G15020
|
unknown |
0.373 |
-0.106 |
| 198 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
0.372 |
0.024 |
| 199 |
252083_at |
AT3G51960
|
ATBZIP24, basic leucine zipper 24, BZIP24, basic leucine zipper 24 |
0.372 |
-0.021 |
| 200 |
244921_s_at |
ATMG01000
|
ORF114 |
0.371 |
-0.029 |
| 201 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.370 |
0.044 |
| 202 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.369 |
0.109 |
| 203 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.369 |
0.117 |
| 204 |
247686_at |
AT5G59700
|
[Protein kinase superfamily protein] |
0.368 |
-0.009 |
| 205 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.368 |
0.039 |
| 206 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.366 |
-0.009 |
| 207 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.366 |
0.050 |
| 208 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
0.365 |
0.032 |
| 209 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
0.362 |
-0.022 |
| 210 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.361 |
0.013 |
| 211 |
254263_at |
AT4G23493
|
unknown |
0.361 |
0.022 |
| 212 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.359 |
0.026 |
| 213 |
257334_at |
ATMG01370
|
ORF111D |
0.357 |
-0.026 |
| 214 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.356 |
-0.074 |
| 215 |
245194_at |
AT1G67820
|
[Protein phosphatase 2C family protein] |
0.355 |
0.057 |
| 216 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.355 |
0.029 |
| 217 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
0.355 |
0.013 |
| 218 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.355 |
0.157 |
| 219 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.355 |
0.040 |
| 220 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.354 |
0.071 |
| 221 |
246607_at |
AT5G35370
|
[S-locus lectin protein kinase family protein] |
0.352 |
0.152 |
| 222 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.352 |
-0.032 |
| 223 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.351 |
0.111 |
| 224 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.351 |
0.275 |
| 225 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.350 |
-0.004 |
| 226 |
245353_at |
AT4G16000
|
unknown |
0.350 |
0.011 |
| 227 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.350 |
-0.070 |
| 228 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
0.350 |
0.008 |
| 229 |
249797_at |
AT5G23750
|
[Remorin family protein] |
0.350 |
-0.065 |
| 230 |
262664_at |
AT1G13970
|
unknown |
0.348 |
-0.077 |
| 231 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.348 |
-0.018 |
| 232 |
264942_at |
AT1G67340
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.347 |
0.018 |
| 233 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.347 |
0.078 |
| 234 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.346 |
0.136 |
| 235 |
247290_at |
AT5G64450
|
unknown |
0.344 |
-0.059 |
| 236 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.344 |
0.016 |
| 237 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
0.343 |
0.006 |
| 238 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.343 |
0.221 |
| 239 |
251086_at |
AT5G01450
|
APD2, ABERRANT POLLEN DEVELOPMENT 2 |
0.343 |
0.015 |
| 240 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.342 |
0.013 |
| 241 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.341 |
0.022 |
| 242 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.340 |
0.037 |
| 243 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.339 |
-0.032 |
| 244 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.338 |
0.032 |
| 245 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.337 |
-0.170 |
| 246 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.337 |
0.047 |
| 247 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.337 |
0.136 |
| 248 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.337 |
0.113 |
| 249 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.335 |
0.149 |
| 250 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.335 |
0.201 |
| 251 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.334 |
0.172 |
| 252 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.334 |
0.173 |
| 253 |
264537_at |
AT1G55610
|
BRL1, BRI1 like |
0.334 |
0.043 |
| 254 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.334 |
-0.042 |
| 255 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.333 |
-0.035 |
| 256 |
250028_at |
AT5G18130
|
unknown |
0.333 |
0.095 |
| 257 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.332 |
0.317 |
| 258 |
263182_at |
AT1G05575
|
unknown |
0.331 |
0.213 |
| 259 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
0.330 |
-0.063 |
| 260 |
260243_at |
AT1G63720
|
unknown |
0.330 |
0.059 |
| 261 |
256210_at |
AT1G50950
|
[INVOLVED IN: cell redox homeostasis] |
0.329 |
0.055 |
| 262 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.329 |
-0.020 |
| 263 |
258103_at |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
0.328 |
0.066 |
| 264 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.328 |
0.049 |
| 265 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
0.327 |
-0.138 |
| 266 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.326 |
-0.044 |
| 267 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
0.326 |
-0.009 |
| 268 |
257074_at |
AT3G19660
|
unknown |
0.323 |
0.093 |
| 269 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.323 |
0.001 |
| 270 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.323 |
0.356 |
| 271 |
248874_at |
AT5G46460
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.323 |
0.011 |
| 272 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.323 |
0.250 |
| 273 |
250141_at |
AT5G14640
|
ATSK13, SHAGGY-LIKE KINASE 13, SK13, shaggy-like kinase 13 |
0.322 |
-0.010 |
| 274 |
257321_at |
ATMG01130
|
ORF106F |
0.322 |
0.025 |
| 275 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
0.322 |
0.216 |
| 276 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.322 |
-0.002 |
| 277 |
263586_at |
AT2G25350
|
[Phox (PX) domain-containing protein] |
0.321 |
0.035 |
| 278 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.321 |
-0.083 |
| 279 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.320 |
0.045 |
| 280 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
0.320 |
0.032 |
| 281 |
249403_at |
AT5G40270
|
[HD domain-containing metal-dependent phosphohydrolase family protein] |
0.320 |
-0.033 |
| 282 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.319 |
0.198 |
| 283 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.317 |
0.188 |
| 284 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.317 |
0.126 |
| 285 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.317 |
0.054 |
| 286 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.316 |
-0.019 |
| 287 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.314 |
0.264 |
| 288 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.313 |
0.053 |
| 289 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.313 |
0.086 |
| 290 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.313 |
0.035 |
| 291 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
0.313 |
-0.043 |
| 292 |
247193_at |
AT5G65380
|
[MATE efflux family protein] |
0.313 |
0.020 |
| 293 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
0.312 |
0.009 |
| 294 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.312 |
0.148 |
| 295 |
246909_at |
AT5G25770
|
[alpha/beta-Hydrolases superfamily protein] |
0.312 |
0.039 |
| 296 |
261535_at |
AT1G01725
|
unknown |
0.311 |
-0.008 |
| 297 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.311 |
0.071 |
| 298 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
0.311 |
-0.067 |
| 299 |
255243_at |
AT4G05590
|
unknown |
0.310 |
0.068 |
| 300 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.310 |
0.006 |
| 301 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-1.466 |
-0.121 |
| 302 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.201 |
-0.110 |
| 303 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.070 |
-0.155 |
| 304 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-1.027 |
0.006 |
| 305 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-1.012 |
0.016 |
| 306 |
261684_at |
AT1G47400
|
unknown |
-1.010 |
0.096 |
| 307 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-1.007 |
-0.050 |
| 308 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.980 |
-0.044 |
| 309 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.958 |
-0.067 |
| 310 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.958 |
-0.112 |
| 311 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.939 |
0.008 |
| 312 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.928 |
-0.035 |
| 313 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
-0.922 |
0.005 |
| 314 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-0.908 |
-0.065 |
| 315 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.905 |
-0.072 |
| 316 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
-0.885 |
-0.079 |
| 317 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.881 |
-0.049 |
| 318 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.871 |
-0.078 |
| 319 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.854 |
-0.168 |
| 320 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.842 |
-0.110 |
| 321 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.818 |
-0.103 |
| 322 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.816 |
-0.002 |
| 323 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.815 |
-0.084 |
| 324 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.813 |
-0.187 |
| 325 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.801 |
-0.250 |
| 326 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.798 |
0.017 |
| 327 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.793 |
-0.184 |
| 328 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.788 |
-0.004 |
| 329 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.781 |
-0.121 |
| 330 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.775 |
-0.216 |
| 331 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.772 |
-0.029 |
| 332 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.772 |
-0.169 |
| 333 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.765 |
-0.046 |
| 334 |
248139_at |
AT5G54970
|
unknown |
-0.758 |
-0.072 |
| 335 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.757 |
-0.195 |
| 336 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.755 |
-0.118 |
| 337 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.749 |
-0.111 |
| 338 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.746 |
-0.099 |
| 339 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.745 |
-0.001 |
| 340 |
262399_at |
AT1G49500
|
unknown |
-0.744 |
-0.041 |
| 341 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.744 |
-0.117 |
| 342 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.732 |
-0.017 |
| 343 |
251142_at |
AT5G01015
|
unknown |
-0.725 |
-0.153 |
| 344 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.723 |
-0.066 |
| 345 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.723 |
-0.173 |
| 346 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.721 |
-0.102 |
| 347 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.712 |
-0.020 |
| 348 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.711 |
-0.107 |
| 349 |
252361_at |
AT3G48490
|
unknown |
-0.710 |
-0.060 |
| 350 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.707 |
-0.088 |
| 351 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.698 |
-0.084 |
| 352 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.697 |
-0.121 |
| 353 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
-0.696 |
0.112 |
| 354 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.695 |
0.000 |
| 355 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.690 |
-0.052 |
| 356 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.689 |
-0.113 |
| 357 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.684 |
-0.151 |
| 358 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.679 |
0.030 |
| 359 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.677 |
-0.136 |
| 360 |
258386_at |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.675 |
-0.060 |
| 361 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.670 |
-0.100 |
| 362 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.670 |
-0.146 |
| 363 |
260877_at |
AT1G21500
|
unknown |
-0.669 |
-0.042 |
| 364 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.667 |
-0.071 |
| 365 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.666 |
-0.068 |
| 366 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.666 |
-0.137 |
| 367 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.663 |
-0.093 |
| 368 |
261203_at |
AT1G12845
|
unknown |
-0.662 |
-0.072 |
| 369 |
249167_at |
AT5G42860
|
unknown |
-0.657 |
-0.043 |
| 370 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.657 |
-0.023 |
| 371 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.657 |
0.007 |
| 372 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
-0.654 |
-0.122 |
| 373 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.653 |
-0.082 |
| 374 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.650 |
0.086 |
| 375 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.650 |
-0.041 |
| 376 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.649 |
-0.038 |
| 377 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.649 |
-0.159 |
| 378 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.648 |
-0.128 |
| 379 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.644 |
-0.185 |
| 380 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.643 |
-0.073 |
| 381 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.642 |
-0.214 |
| 382 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.641 |
-0.119 |
| 383 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.640 |
-0.114 |
| 384 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.634 |
-0.170 |
| 385 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.628 |
-0.026 |
| 386 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.623 |
-0.071 |
| 387 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-0.614 |
-0.096 |
| 388 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
-0.610 |
-0.098 |
| 389 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.607 |
-0.035 |
| 390 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.607 |
-0.144 |
| 391 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.606 |
-0.063 |
| 392 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.605 |
-0.031 |
| 393 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.603 |
-0.064 |
| 394 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.597 |
-0.064 |
| 395 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.596 |
0.074 |
| 396 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.593 |
-0.093 |
| 397 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.592 |
-0.078 |
| 398 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.592 |
-0.052 |
| 399 |
253305_at |
AT4G33666
|
unknown |
-0.591 |
-0.054 |
| 400 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.588 |
-0.096 |
| 401 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.588 |
-0.102 |
| 402 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.587 |
-0.111 |
| 403 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.586 |
-0.037 |
| 404 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
-0.584 |
-0.069 |
| 405 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.582 |
-0.035 |
| 406 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.582 |
-0.081 |
| 407 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.582 |
-0.117 |
| 408 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
-0.576 |
-0.064 |
| 409 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.572 |
-0.052 |
| 410 |
253875_at |
AT4G27520
|
AtENODL2, ENODL2, early nodulin-like protein 2 |
-0.571 |
-0.036 |
| 411 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.571 |
-0.170 |
| 412 |
261921_at |
AT1G65900
|
unknown |
-0.571 |
-0.197 |
| 413 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.568 |
-0.131 |
| 414 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.568 |
-0.161 |
| 415 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.567 |
-0.014 |
| 416 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.564 |
0.060 |
| 417 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.563 |
0.021 |
| 418 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.562 |
-0.071 |
| 419 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.562 |
-0.069 |
| 420 |
245112_at |
AT2G41540
|
GPDHC1 |
-0.560 |
0.085 |
| 421 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.557 |
-0.097 |
| 422 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.553 |
-0.218 |
| 423 |
258641_at |
AT3G08030
|
unknown |
-0.552 |
-0.085 |
| 424 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.552 |
-0.049 |
| 425 |
264343_at |
AT1G11850
|
unknown |
-0.549 |
-0.122 |
| 426 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.547 |
-0.071 |
| 427 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.544 |
-0.120 |
| 428 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.544 |
-0.074 |
| 429 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.540 |
-0.172 |
| 430 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.539 |
-0.065 |
| 431 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.539 |
-0.060 |
| 432 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.539 |
0.001 |
| 433 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.537 |
-0.004 |
| 434 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.535 |
0.040 |
| 435 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.535 |
-0.110 |
| 436 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.535 |
-0.087 |
| 437 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.534 |
-0.108 |
| 438 |
248509_at |
AT5G50335
|
unknown |
-0.533 |
-0.061 |
| 439 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.532 |
-0.096 |
| 440 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.531 |
-0.011 |
| 441 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.531 |
-0.026 |
| 442 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.528 |
0.001 |
| 443 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.526 |
0.015 |
| 444 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.522 |
-0.129 |
| 445 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.517 |
-0.084 |
| 446 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
-0.517 |
-0.077 |
| 447 |
256021_at |
AT1G58270
|
ZW9 |
-0.514 |
0.004 |
| 448 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
-0.511 |
0.019 |
| 449 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.507 |
-0.105 |
| 450 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.507 |
-0.073 |
| 451 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.506 |
-0.132 |
| 452 |
248861_at |
AT5G46700
|
TET1, TETRASPANIN 1, TRN2, TORNADO 2 |
-0.506 |
-0.026 |
| 453 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.505 |
-0.116 |
| 454 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.504 |
-0.012 |
| 455 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.504 |
-0.042 |
| 456 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.504 |
-0.126 |
| 457 |
251852_at |
AT3G54750
|
unknown |
-0.501 |
-0.032 |
| 458 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.500 |
-0.041 |
| 459 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.500 |
-0.049 |
| 460 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
-0.498 |
0.022 |
| 461 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.495 |
-0.115 |
| 462 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.494 |
-0.042 |
| 463 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.492 |
-0.020 |
| 464 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.492 |
0.030 |
| 465 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.492 |
-0.022 |
| 466 |
250828_at |
AT5G05250
|
unknown |
-0.491 |
0.084 |
| 467 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.490 |
-0.102 |
| 468 |
261639_at |
AT1G50010
|
TUA2, tubulin alpha-2 chain |
-0.490 |
0.005 |
| 469 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.490 |
-0.103 |
| 470 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.490 |
-0.091 |
| 471 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.490 |
-0.102 |
| 472 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
-0.488 |
-0.046 |
| 473 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.487 |
-0.021 |
| 474 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.487 |
-0.057 |
| 475 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.486 |
0.146 |
| 476 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.486 |
-0.013 |
| 477 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.485 |
-0.069 |
| 478 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.482 |
0.035 |
| 479 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.481 |
-0.045 |
| 480 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.481 |
-0.110 |
| 481 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.480 |
-0.040 |
| 482 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.480 |
-0.075 |
| 483 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.479 |
-0.114 |
| 484 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.476 |
0.038 |
| 485 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.475 |
0.012 |
| 486 |
265183_at |
AT1G23750
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.474 |
-0.094 |
| 487 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
-0.474 |
0.037 |
| 488 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.474 |
-0.101 |
| 489 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.473 |
-0.056 |
| 490 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.473 |
-0.062 |
| 491 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
-0.472 |
-0.066 |
| 492 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.472 |
-0.349 |
| 493 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.470 |
-0.078 |
| 494 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.470 |
-0.179 |
| 495 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.469 |
-0.197 |
| 496 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.468 |
0.022 |
| 497 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.468 |
0.057 |
| 498 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.463 |
-0.079 |
| 499 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.462 |
0.021 |
| 500 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.462 |
-0.028 |
| 501 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.462 |
0.023 |
| 502 |
263640_at |
AT2G25270
|
unknown |
-0.462 |
-0.052 |
| 503 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.460 |
-0.038 |
| 504 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.460 |
-0.160 |
| 505 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.460 |
-0.017 |
| 506 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.459 |
-0.099 |
| 507 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.458 |
-0.106 |
| 508 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.458 |
0.018 |
| 509 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.456 |
-0.098 |
| 510 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.453 |
-0.048 |
| 511 |
251036_at |
AT5G02160
|
unknown |
-0.452 |
-0.043 |
| 512 |
249090_at |
AT5G43745
|
unknown |
-0.451 |
-0.064 |
| 513 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.450 |
-0.147 |
| 514 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.450 |
-0.023 |
| 515 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.450 |
-0.010 |
| 516 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.448 |
-0.063 |
| 517 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.447 |
-0.065 |
| 518 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.444 |
-0.092 |
| 519 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.443 |
-0.097 |
| 520 |
250441_at |
AT5G10540
|
TOP2, thimet metalloendopeptidase 2 |
-0.441 |
-0.027 |
| 521 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.441 |
-0.091 |
| 522 |
261492_at |
AT1G14290
|
SBH2, sphingoid base hydroxylase 2 |
-0.440 |
-0.084 |
| 523 |
256096_at |
AT1G13650
|
unknown |
-0.439 |
-0.220 |
| 524 |
253817_at |
AT4G28310
|
unknown |
-0.439 |
-0.120 |
| 525 |
258468_at |
AT3G06070
|
unknown |
-0.438 |
0.000 |
| 526 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.438 |
-0.096 |
| 527 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.438 |
-0.064 |
| 528 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.438 |
-0.173 |
| 529 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.438 |
-0.075 |
| 530 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
-0.437 |
-0.040 |
| 531 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.435 |
-0.097 |
| 532 |
246265_at |
AT1G31860
|
AT-IE, HISN2, HISTIDINE BIOSYNTHESIS 2 |
-0.435 |
-0.016 |
| 533 |
251219_at |
AT3G62390
|
TBL6, TRICHOME BIREFRINGENCE-LIKE 6 |
-0.433 |
-0.113 |
| 534 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.433 |
-0.110 |
| 535 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.432 |
0.181 |
| 536 |
266334_at |
AT2G32380
|
[Transmembrane protein 97, predicted] |
-0.427 |
0.052 |
| 537 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.425 |
-0.171 |
| 538 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
-0.425 |
0.069 |
| 539 |
263535_at |
AT2G24970
|
unknown |
-0.423 |
-0.068 |
| 540 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.423 |
-0.099 |
| 541 |
252215_at |
AT3G50240
|
KICP-02 |
-0.422 |
-0.070 |
| 542 |
259761_at |
AT1G77590
|
LACS9, long chain acyl-CoA synthetase 9 |
-0.422 |
-0.055 |
| 543 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.421 |
-0.018 |
| 544 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.420 |
-0.117 |
| 545 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.419 |
-0.019 |
| 546 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.418 |
-0.061 |
| 547 |
258368_at |
AT3G14240
|
[Subtilase family protein] |
-0.418 |
-0.067 |
| 548 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.417 |
-0.057 |
| 549 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.417 |
-0.067 |
| 550 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.417 |
-0.008 |
| 551 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.416 |
-0.039 |
| 552 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.416 |
-0.048 |
| 553 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.415 |
0.027 |
| 554 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.415 |
-0.085 |
| 555 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.415 |
-0.056 |
| 556 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.414 |
-0.123 |
| 557 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.414 |
-0.134 |
| 558 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.414 |
-0.098 |
| 559 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.414 |
0.129 |
| 560 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.412 |
-0.007 |
| 561 |
245215_at |
AT1G67830
|
ATFXG1, Arabidopsis thaliana alpha-fucosidase 1, FXG1, alpha-fucosidase 1 |
-0.409 |
-0.118 |
| 562 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.409 |
-0.050 |
| 563 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.408 |
-0.067 |
| 564 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.407 |
-0.098 |
| 565 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.407 |
-0.113 |
| 566 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
-0.407 |
-0.145 |
| 567 |
258123_at |
AT3G18040
|
MPK9, MAP kinase 9 |
-0.406 |
-0.055 |
| 568 |
251736_at |
AT3G56130
|
[biotin/lipoyl attachment domain-containing protein] |
-0.404 |
-0.108 |
| 569 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.403 |
-0.086 |
| 570 |
260944_at |
AT1G45130
|
AtBGAL5, BGAL5, beta-galactosidase 5 |
-0.402 |
-0.089 |
| 571 |
246200_at |
AT4G37240
|
unknown |
-0.401 |
-0.129 |
| 572 |
250366_at |
AT5G11420
|
unknown |
-0.400 |
-0.090 |
| 573 |
258156_at |
AT3G18050
|
unknown |
-0.400 |
-0.096 |
| 574 |
265824_at |
AT2G35650
|
ATCSLA07, cellulose synthase like, ATCSLA7, CSLA07, CSLA07, cellulose synthase like, CSLA7, CELLULOSE SYNTHASE LIKE A7 |
-0.400 |
-0.002 |
| 575 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.399 |
-0.082 |
| 576 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.399 |
-0.104 |
| 577 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.399 |
-0.014 |
| 578 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.398 |
-0.106 |
| 579 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.396 |
-0.105 |
| 580 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.396 |
0.001 |
| 581 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.396 |
-0.137 |
| 582 |
246792_at |
AT5G27290
|
unknown |
-0.395 |
-0.045 |
| 583 |
246269_at |
AT4G37110
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
-0.394 |
-0.063 |
| 584 |
245368_at |
AT4G15510
|
PPD1, PsbP-Domain Protein1 |
-0.393 |
-0.082 |
| 585 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.393 |
0.051 |
| 586 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.393 |
-0.028 |
| 587 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.393 |
-0.119 |
| 588 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.393 |
-0.028 |
| 589 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.392 |
-0.068 |
| 590 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.392 |
-0.134 |
| 591 |
257219_at |
AT3G14930
|
HEME1 |
-0.392 |
-0.046 |
| 592 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.392 |
-0.013 |
| 593 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.391 |
-0.067 |
| 594 |
263202_at |
AT1G05630
|
5PTASE13, inositol-polyphosphate 5-phosphatase 13, AT5PTASE13, Arabidopsis thaliana inositol-polyphosphate 5-phosphatase 13 |
-0.391 |
0.021 |
| 595 |
258462_at |
AT3G17350
|
unknown |
-0.391 |
-0.004 |
| 596 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.391 |
-0.063 |
| 597 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.390 |
-0.203 |
| 598 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.389 |
-0.083 |
| 599 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.388 |
0.182 |
| 600 |
261223_at |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
-0.387 |
-0.124 |