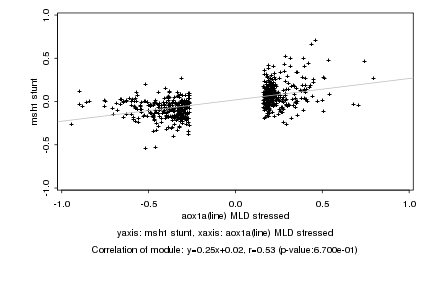
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
aox1a(line) MLD stressed |
Log2 signal ratio
msh1 stunt |
| 1 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.793 |
0.277 |
| 2 |
264580_at |
AT1G05340
|
unknown |
0.738 |
0.465 |
| 3 |
253595_at |
AT4G30830
|
unknown |
0.707 |
-0.039 |
| 4 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.679 |
-0.030 |
| 5 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.536 |
0.083 |
| 6 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.532 |
0.485 |
| 7 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.508 |
0.276 |
| 8 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.505 |
0.288 |
| 9 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
0.502 |
-0.109 |
| 10 |
245799_at |
AT1G45616
|
AtRLP6, receptor like protein 6, RLP6, receptor like protein 6 |
0.500 |
0.020 |
| 11 |
251012_at |
AT5G02580
|
unknown |
0.468 |
0.006 |
| 12 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.457 |
0.716 |
| 13 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.449 |
0.258 |
| 14 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.444 |
0.226 |
| 15 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.438 |
0.063 |
| 16 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.433 |
0.661 |
| 17 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.427 |
0.188 |
| 18 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.422 |
0.163 |
| 19 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.418 |
0.441 |
| 20 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.410 |
0.002 |
| 21 |
248505_at |
AT5G50360
|
unknown |
0.408 |
0.145 |
| 22 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.405 |
0.033 |
| 23 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.402 |
0.187 |
| 24 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.401 |
0.277 |
| 25 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.401 |
0.109 |
| 26 |
266640_at |
AT2G35585
|
unknown |
0.400 |
0.027 |
| 27 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.397 |
0.148 |
| 28 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.396 |
0.414 |
| 29 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.395 |
0.129 |
| 30 |
246703_at |
AT5G28080
|
WNK9 |
0.395 |
0.036 |
| 31 |
258209_at |
AT3G14060
|
unknown |
0.391 |
0.498 |
| 32 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.390 |
-0.095 |
| 33 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.382 |
0.286 |
| 34 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.381 |
0.189 |
| 35 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.379 |
0.124 |
| 36 |
265954_at |
AT2G37260
|
ATWRKY44, DSL1, DR. STRANGELOVE 1, TTG2, TRANSPARENT TESTA GLABRA 2, WRKY44 |
0.376 |
0.035 |
| 37 |
250158_at |
AT5G15190
|
unknown |
0.371 |
0.134 |
| 38 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.357 |
0.347 |
| 39 |
260553_at |
AT2G41800
|
unknown |
0.357 |
0.002 |
| 40 |
262601_at |
AT1G15310
|
ATHSRP54A, signal recognition particle 54 kDa subunit, SRP54-1, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT |
0.354 |
-0.158 |
| 41 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.351 |
-0.014 |
| 42 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.350 |
0.336 |
| 43 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.350 |
0.005 |
| 44 |
260005_at |
AT1G67920
|
unknown |
0.346 |
0.158 |
| 45 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
0.345 |
0.038 |
| 46 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.345 |
-0.064 |
| 47 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.341 |
-0.003 |
| 48 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.340 |
0.181 |
| 49 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.336 |
0.091 |
| 50 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.333 |
-0.037 |
| 51 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.330 |
0.192 |
| 52 |
256789_at |
AT3G13672
|
SINA2 |
0.327 |
0.028 |
| 53 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.320 |
0.182 |
| 54 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.318 |
-0.185 |
| 55 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.316 |
0.415 |
| 56 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.315 |
0.506 |
| 57 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
0.311 |
-0.045 |
| 58 |
257866_at |
AT3G17770
|
[Dihydroxyacetone kinase] |
0.303 |
0.217 |
| 59 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.302 |
0.147 |
| 60 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.301 |
-0.023 |
| 61 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.297 |
0.296 |
| 62 |
263881_at |
AT2G21820
|
unknown |
0.296 |
-0.049 |
| 63 |
250388_at |
AT5G11310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.293 |
-0.040 |
| 64 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.292 |
0.031 |
| 65 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.292 |
-0.262 |
| 66 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.289 |
-0.026 |
| 67 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.289 |
0.147 |
| 68 |
249622_at |
AT5G37550
|
unknown |
0.289 |
-0.030 |
| 69 |
256603_at |
AT3G28270
|
unknown |
0.288 |
0.071 |
| 70 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.285 |
0.524 |
| 71 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.285 |
0.241 |
| 72 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.285 |
0.100 |
| 73 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.283 |
0.023 |
| 74 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.282 |
0.432 |
| 75 |
249454_at |
AT5G39520
|
unknown |
0.279 |
0.358 |
| 76 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.276 |
0.057 |
| 77 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.274 |
-0.237 |
| 78 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.272 |
-0.087 |
| 79 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.272 |
0.130 |
| 80 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
0.271 |
-0.002 |
| 81 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.270 |
-0.117 |
| 82 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.268 |
-0.061 |
| 83 |
252606_at |
AT3G45010
|
scpl48, serine carboxypeptidase-like 48 |
0.268 |
-0.079 |
| 84 |
256442_at |
AT3G10930
|
unknown |
0.268 |
0.005 |
| 85 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.266 |
0.030 |
| 86 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.265 |
-0.118 |
| 87 |
262201_at |
AT2G01120
|
ATORC4, ORIGIN RECOGNITION COMPLEX SUBUNIT 4, ORC4, origin recognition complex subunit 4 |
0.264 |
-0.059 |
| 88 |
252400_at |
AT3G48020
|
unknown |
0.257 |
0.050 |
| 89 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.256 |
0.076 |
| 90 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.254 |
-0.113 |
| 91 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.254 |
0.338 |
| 92 |
264287_at |
AT1G61930
|
unknown |
0.253 |
-0.091 |
| 93 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
0.253 |
0.275 |
| 94 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.252 |
0.003 |
| 95 |
260163_at |
AT1G79900
|
ATMBAC2, RABIDOPSIS MITOCHONDRIAL BASIC AMINO ACID CARRIER 2, BAC2 |
0.251 |
0.019 |
| 96 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.246 |
0.048 |
| 97 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.244 |
0.015 |
| 98 |
247519_at |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
0.244 |
0.088 |
| 99 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.244 |
-0.149 |
| 100 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.243 |
0.082 |
| 101 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
0.243 |
0.106 |
| 102 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.241 |
0.093 |
| 103 |
259901_at |
AT1G74180
|
AtRLP14, receptor like protein 14, RLP14, receptor like protein 14 |
0.238 |
0.027 |
| 104 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.236 |
-0.016 |
| 105 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.235 |
0.174 |
| 106 |
254853_at |
AT4G12080
|
AHL1, AT-hook motif nuclear-localized protein 1, ATAHL1 |
0.235 |
0.018 |
| 107 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.235 |
-0.035 |
| 108 |
258782_at |
AT3G11750
|
FOLB1 |
0.235 |
0.032 |
| 109 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.234 |
0.050 |
| 110 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.233 |
0.015 |
| 111 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.231 |
-0.022 |
| 112 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.230 |
0.073 |
| 113 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.229 |
0.092 |
| 114 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.228 |
0.196 |
| 115 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.227 |
0.263 |
| 116 |
256887_at |
AT3G15150
|
ATMMS21, A. THALIANA METHYL METHANE SULFONATE SENSITIVITY 21, HPY2, HIGH PLOIDY2, MMS21, METHYL METHANE SULFONATE SENSITIVITY 2 |
0.227 |
0.107 |
| 117 |
265059_at |
AT1G52080
|
AR791 |
0.226 |
0.017 |
| 118 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.225 |
0.201 |
| 119 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.224 |
0.080 |
| 120 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
0.224 |
-0.162 |
| 121 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.222 |
0.326 |
| 122 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.222 |
0.286 |
| 123 |
257395_at |
AT2G15630
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.222 |
0.151 |
| 124 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
0.220 |
0.134 |
| 125 |
267178_at |
AT2G37750
|
unknown |
0.220 |
0.121 |
| 126 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.220 |
-0.028 |
| 127 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.220 |
0.067 |
| 128 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
0.219 |
0.175 |
| 129 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.217 |
0.226 |
| 130 |
247290_at |
AT5G64450
|
unknown |
0.217 |
-0.049 |
| 131 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.217 |
0.154 |
| 132 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.217 |
0.157 |
| 133 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.217 |
0.405 |
| 134 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.216 |
0.132 |
| 135 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.215 |
0.095 |
| 136 |
258103_at |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
0.214 |
-0.022 |
| 137 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.214 |
0.039 |
| 138 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.214 |
0.139 |
| 139 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.213 |
0.015 |
| 140 |
259881_at |
AT1G76820
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
0.213 |
0.007 |
| 141 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.213 |
0.035 |
| 142 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.213 |
0.016 |
| 143 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.212 |
0.183 |
| 144 |
264818_at |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
0.212 |
0.083 |
| 145 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.212 |
-0.042 |
| 146 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.212 |
0.204 |
| 147 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.212 |
0.266 |
| 148 |
264294_at |
AT1G78750
|
[F-box/RNI-like superfamily protein] |
0.210 |
0.118 |
| 149 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.210 |
0.162 |
| 150 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
0.209 |
0.182 |
| 151 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.209 |
0.199 |
| 152 |
265276_at |
AT2G28400
|
unknown |
0.209 |
0.189 |
| 153 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
0.209 |
-0.124 |
| 154 |
250825_at |
AT5G05210
|
[Surfeit locus protein 6] |
0.208 |
-0.011 |
| 155 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.208 |
0.143 |
| 156 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.208 |
0.139 |
| 157 |
246125_at |
AT5G19875
|
unknown |
0.207 |
0.036 |
| 158 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.207 |
0.100 |
| 159 |
258411_at |
AT3G17280
|
[F-box and associated interaction domains-containing protein] |
0.207 |
-0.042 |
| 160 |
258118_at |
AT3G14710
|
[RNI-like superfamily protein] |
0.206 |
0.047 |
| 161 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.206 |
0.082 |
| 162 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.205 |
0.068 |
| 163 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.205 |
0.028 |
| 164 |
266064_at |
AT2G18780
|
[F-box and associated interaction domains-containing protein] |
0.205 |
0.024 |
| 165 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.205 |
0.116 |
| 166 |
262478_at |
AT1G11170
|
unknown |
0.205 |
0.012 |
| 167 |
255494_at |
AT4G02710
|
NET1C, Networked 1C |
0.204 |
0.009 |
| 168 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
0.204 |
0.043 |
| 169 |
252990_at |
AT4G38440
|
IYO, MINIYO |
0.204 |
0.174 |
| 170 |
248239_at |
AT5G53920
|
[ribosomal protein L11 methyltransferase-related] |
0.204 |
0.043 |
| 171 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.204 |
0.312 |
| 172 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.203 |
0.105 |
| 173 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.203 |
0.031 |
| 174 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.203 |
0.017 |
| 175 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.203 |
0.079 |
| 176 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.202 |
0.066 |
| 177 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
0.202 |
0.225 |
| 178 |
261972_at |
AT1G64600
|
[methyltransferases] |
0.200 |
0.201 |
| 179 |
256861_at |
AT3G23920
|
AtBAM1, BAM1, beta-amylase 1, BMY7, BETA-AMYLASE 7, TR-BAMY |
0.200 |
0.116 |
| 180 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.199 |
0.158 |
| 181 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.199 |
0.017 |
| 182 |
252470_at |
AT3G46930
|
[Protein kinase superfamily protein] |
0.199 |
-0.031 |
| 183 |
262625_at |
AT1G06440
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.198 |
0.013 |
| 184 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.198 |
0.231 |
| 185 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.198 |
0.008 |
| 186 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.197 |
0.006 |
| 187 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.197 |
0.270 |
| 188 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.197 |
0.156 |
| 189 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.197 |
0.090 |
| 190 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.196 |
0.227 |
| 191 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
0.196 |
-0.028 |
| 192 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
0.196 |
-0.023 |
| 193 |
254421_at |
AT4G21550
|
VAL3, VP1/ABI3-like 3 |
0.195 |
-0.127 |
| 194 |
259489_at |
AT1G15790
|
unknown |
0.195 |
-0.006 |
| 195 |
258248_at |
AT3G29140
|
unknown |
0.195 |
-0.007 |
| 196 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.195 |
0.202 |
| 197 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.195 |
-0.084 |
| 198 |
266775_at |
AT2G29065
|
[GRAS family transcription factor] |
0.195 |
-0.093 |
| 199 |
255337_at |
AT4G04450
|
AtWRKY42, WRKY42 |
0.195 |
-0.076 |
| 200 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.194 |
0.107 |
| 201 |
245905_at |
AT5G11090
|
[serine-rich protein-related] |
0.194 |
-0.012 |
| 202 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
0.194 |
0.235 |
| 203 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.193 |
0.193 |
| 204 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.192 |
0.063 |
| 205 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.192 |
0.052 |
| 206 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.191 |
0.047 |
| 207 |
252469_at |
AT3G46920
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
0.191 |
0.195 |
| 208 |
247528_at |
AT5G61460
|
ATRAD18, MIM, hypersensitive to MMS, irradiation and MMC, SMC6B, STRUCTURAL MAINTENANCE OF CHROMOSOMES 6B |
0.191 |
0.000 |
| 209 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.190 |
0.084 |
| 210 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.190 |
0.420 |
| 211 |
267519_at |
AT2G30470
|
HSI2, high-level expression of sugar-inducible gene 2, VAL1, VP1/ABI3-LIKE 1 |
0.190 |
0.147 |
| 212 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.190 |
-0.061 |
| 213 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
0.190 |
0.104 |
| 214 |
261890_at |
AT1G80970
|
[XH domain-containing protein] |
0.189 |
-0.008 |
| 215 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.189 |
0.071 |
| 216 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
0.189 |
-0.057 |
| 217 |
244903_at |
ATMG00660
|
ORF149 |
0.188 |
0.071 |
| 218 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
0.188 |
0.138 |
| 219 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
0.188 |
-0.105 |
| 220 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
0.188 |
-0.166 |
| 221 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.188 |
0.194 |
| 222 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.187 |
-0.005 |
| 223 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.187 |
0.048 |
| 224 |
250771_at |
AT5G05400
|
[LRR and NB-ARC domains-containing disease resistance protein] |
0.187 |
0.023 |
| 225 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.187 |
0.387 |
| 226 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
0.186 |
-0.163 |
| 227 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.186 |
-0.017 |
| 228 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.186 |
0.249 |
| 229 |
249252_at |
AT5G42010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.185 |
0.133 |
| 230 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.185 |
0.082 |
| 231 |
256210_at |
AT1G50950
|
[INVOLVED IN: cell redox homeostasis] |
0.184 |
0.007 |
| 232 |
260062_at |
AT1G73710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.184 |
0.130 |
| 233 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.182 |
-0.131 |
| 234 |
246614_at |
AT5G35410
|
ATSOS2, SALT OVERLY SENSITIVE 2, CIPK24, CBL-INTERACTING PROTEIN KINASE 24, SNRK3.11, SNF1-RELATED PROTEIN KINASE 3.11, SOS2, SALT OVERLY SENSITIVE 2 |
0.182 |
-0.025 |
| 235 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
0.182 |
0.163 |
| 236 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.182 |
0.287 |
| 237 |
264604_at |
AT1G04650
|
unknown |
0.182 |
0.067 |
| 238 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.181 |
0.038 |
| 239 |
245147_at |
AT2G45280
|
ATRAD51C, RAS associated with diabetes protein 51C, RAD51C, RAS associated with diabetes protein 51C |
0.181 |
-0.023 |
| 240 |
256276_at |
AT3G12070
|
AtRGTB2, RAB geranylgeranyl transferase beta subunit 2, RGTB2, RAB geranylgeranyl transferase beta subunit 2 |
0.181 |
0.062 |
| 241 |
255647_at |
AT4G00900
|
ATECA2, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 2, ECA2, ER-type Ca2+-ATPase 2 |
0.181 |
0.081 |
| 242 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.181 |
0.119 |
| 243 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.180 |
-0.075 |
| 244 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.180 |
-0.055 |
| 245 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.179 |
0.298 |
| 246 |
253965_at |
AT4G26490
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.179 |
-0.153 |
| 247 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.179 |
0.139 |
| 248 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.179 |
0.018 |
| 249 |
255481_at |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
0.178 |
0.039 |
| 250 |
248278_at |
AT5G52890
|
[AT hook motif-containing protein] |
0.178 |
0.134 |
| 251 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.178 |
-0.081 |
| 252 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.178 |
0.110 |
| 253 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.178 |
-0.092 |
| 254 |
256377_at |
AT1G66650
|
[Protein with RING/U-box and TRAF-like domains] |
0.178 |
0.067 |
| 255 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.177 |
0.152 |
| 256 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
0.177 |
-0.053 |
| 257 |
260733_at |
AT1G17640
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.177 |
0.231 |
| 258 |
249797_at |
AT5G23750
|
[Remorin family protein] |
0.177 |
-0.044 |
| 259 |
261945_at |
AT1G64530
|
NLP6, NIN-like protein 6 |
0.177 |
0.038 |
| 260 |
264483_at |
AT1G77230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.176 |
-0.012 |
| 261 |
262074_at |
AT1G59670
|
ATGSTU15, glutathione S-transferase TAU 15, GSTU15, glutathione S-transferase TAU 15 |
0.176 |
0.016 |
| 262 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.176 |
0.088 |
| 263 |
259856_at |
AT1G68440
|
unknown |
0.176 |
0.323 |
| 264 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
0.175 |
0.197 |
| 265 |
259922_at |
AT1G72770
|
AtHAB1, HAB1, HYPERSENSITIVE TO ABA1 |
0.175 |
0.143 |
| 266 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
0.175 |
0.044 |
| 267 |
254020_at |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
0.174 |
0.171 |
| 268 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.173 |
-0.180 |
| 269 |
252986_at |
AT4G38380
|
[MATE efflux family protein] |
0.172 |
0.082 |
| 270 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.171 |
0.123 |
| 271 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.171 |
-0.181 |
| 272 |
250769_at |
AT5G05680
|
EMB2789, EMBRYO DEFECTIVE 2789, MOS7, MODIFIER OF SNC1,7 |
0.171 |
0.107 |
| 273 |
264749_at |
AT1G22830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.171 |
0.025 |
| 274 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.171 |
-0.077 |
| 275 |
264456_at |
AT1G10390
|
[Nucleoporin autopeptidase] |
0.171 |
0.059 |
| 276 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.168 |
0.153 |
| 277 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
0.168 |
-0.081 |
| 278 |
251957_at |
AT3G53690
|
[RING/U-box superfamily protein] |
0.168 |
0.127 |
| 279 |
265907_at |
AT2G25650
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.167 |
-0.187 |
| 280 |
260160_at |
AT1G79880
|
AtLa2, La2, La protein 2 |
0.167 |
0.057 |
| 281 |
254263_at |
AT4G23493
|
unknown |
0.167 |
-0.096 |
| 282 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
0.166 |
-0.022 |
| 283 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.166 |
0.368 |
| 284 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.165 |
-0.023 |
| 285 |
251177_at |
AT3G63400
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.165 |
0.114 |
| 286 |
260049_at |
AT1G29940
|
NRPA2, nuclear RNA polymerase A2 |
0.165 |
0.238 |
| 287 |
245662_at |
AT1G28190
|
unknown |
0.165 |
-0.005 |
| 288 |
256063_at |
AT1G07130
|
ATSTN1, STN1 |
0.165 |
-0.078 |
| 289 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
0.164 |
0.033 |
| 290 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
0.164 |
0.091 |
| 291 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.164 |
0.314 |
| 292 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.163 |
0.173 |
| 293 |
257073_at |
AT3G19650
|
[cyclin-related] |
0.163 |
0.156 |
| 294 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.161 |
0.018 |
| 295 |
253047_at |
AT4G37295
|
unknown |
0.161 |
-0.012 |
| 296 |
265913_at |
AT2G25625
|
unknown |
0.161 |
-0.015 |
| 297 |
267266_at |
AT2G23150
|
ATNRAMP3, NRAMP3, natural resistance-associated macrophage protein 3 |
0.161 |
0.064 |
| 298 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.161 |
0.185 |
| 299 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
0.160 |
-0.061 |
| 300 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
0.159 |
0.110 |
| 301 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.948 |
-0.264 |
| 302 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.903 |
0.124 |
| 303 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.903 |
-0.026 |
| 304 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.883 |
-0.056 |
| 305 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.859 |
-0.007 |
| 306 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.845 |
0.005 |
| 307 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.759 |
0.021 |
| 308 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.759 |
-0.048 |
| 309 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.749 |
0.005 |
| 310 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.712 |
-0.074 |
| 311 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-0.705 |
-0.139 |
| 312 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.687 |
-0.016 |
| 313 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.680 |
-0.100 |
| 314 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.664 |
-0.025 |
| 315 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.664 |
0.025 |
| 316 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.656 |
0.034 |
| 317 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.650 |
-0.181 |
| 318 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.649 |
-0.002 |
| 319 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.645 |
0.008 |
| 320 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.632 |
-0.141 |
| 321 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.629 |
0.011 |
| 322 |
264341_at |
AT1G70270
|
unknown |
-0.628 |
0.020 |
| 323 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.610 |
0.045 |
| 324 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.601 |
0.005 |
| 325 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.601 |
-0.039 |
| 326 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.599 |
-0.146 |
| 327 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.597 |
0.031 |
| 328 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.588 |
-0.182 |
| 329 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.587 |
-0.201 |
| 330 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.583 |
-0.057 |
| 331 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.583 |
0.007 |
| 332 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.577 |
-0.221 |
| 333 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-0.577 |
-0.061 |
| 334 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.577 |
-0.079 |
| 335 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.577 |
0.037 |
| 336 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.575 |
0.114 |
| 337 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.572 |
0.008 |
| 338 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.565 |
-0.082 |
| 339 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.565 |
0.070 |
| 340 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.563 |
-0.193 |
| 341 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.559 |
-0.242 |
| 342 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.551 |
-0.083 |
| 343 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.551 |
-0.047 |
| 344 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.549 |
-0.044 |
| 345 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.544 |
-0.122 |
| 346 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.542 |
-0.089 |
| 347 |
260877_at |
AT1G21500
|
unknown |
-0.542 |
0.000 |
| 348 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.531 |
-0.171 |
| 349 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.528 |
-0.160 |
| 350 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.528 |
0.006 |
| 351 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.519 |
-0.537 |
| 352 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.519 |
0.004 |
| 353 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
-0.518 |
0.205 |
| 354 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.510 |
0.000 |
| 355 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-0.509 |
-0.168 |
| 356 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.507 |
-0.009 |
| 357 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.504 |
-0.094 |
| 358 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.502 |
-0.041 |
| 359 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.494 |
-0.029 |
| 360 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.490 |
-0.118 |
| 361 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.490 |
-0.047 |
| 362 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.490 |
-0.127 |
| 363 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.488 |
-0.135 |
| 364 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.487 |
-0.044 |
| 365 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.483 |
-0.172 |
| 366 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.476 |
-0.184 |
| 367 |
260112_at |
AT1G63310
|
unknown |
-0.476 |
-0.152 |
| 368 |
248509_at |
AT5G50335
|
unknown |
-0.473 |
-0.341 |
| 369 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.473 |
-0.168 |
| 370 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.472 |
-0.036 |
| 371 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.470 |
-0.137 |
| 372 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.468 |
-0.214 |
| 373 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.468 |
-0.203 |
| 374 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.468 |
-0.013 |
| 375 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.466 |
-0.010 |
| 376 |
249167_at |
AT5G42860
|
unknown |
-0.465 |
-0.529 |
| 377 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.463 |
0.015 |
| 378 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.463 |
-0.093 |
| 379 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.460 |
-0.248 |
| 380 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.459 |
-0.334 |
| 381 |
247474_at |
AT5G62280
|
unknown |
-0.459 |
-0.122 |
| 382 |
260081_at |
AT1G78170
|
unknown |
-0.456 |
-0.014 |
| 383 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.454 |
-0.028 |
| 384 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.448 |
-0.128 |
| 385 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.446 |
-0.069 |
| 386 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.445 |
-0.285 |
| 387 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.443 |
0.107 |
| 388 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.442 |
-0.170 |
| 389 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.442 |
-0.100 |
| 390 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.441 |
-0.036 |
| 391 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.438 |
-0.019 |
| 392 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.430 |
-0.191 |
| 393 |
266865_at |
AT2G29980
|
AtFAD3, FAD3, fatty acid desaturase 3 |
-0.429 |
-0.038 |
| 394 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.428 |
-0.237 |
| 395 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.428 |
-0.074 |
| 396 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.424 |
-0.133 |
| 397 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
-0.418 |
-0.010 |
| 398 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.417 |
-0.113 |
| 399 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.413 |
-0.030 |
| 400 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.411 |
-0.067 |
| 401 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.410 |
-0.040 |
| 402 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.409 |
0.035 |
| 403 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.409 |
-0.119 |
| 404 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.409 |
-0.095 |
| 405 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.408 |
-0.194 |
| 406 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.407 |
0.160 |
| 407 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.402 |
-0.008 |
| 408 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.402 |
-0.134 |
| 409 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.401 |
-0.173 |
| 410 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.401 |
-0.312 |
| 411 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.401 |
-0.154 |
| 412 |
250327_at |
AT5G12050
|
unknown |
-0.400 |
-0.079 |
| 413 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.395 |
0.052 |
| 414 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.394 |
-0.123 |
| 415 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.392 |
-0.116 |
| 416 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
-0.391 |
-0.163 |
| 417 |
249303_at |
AT5G41460
|
unknown |
-0.389 |
-0.294 |
| 418 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.389 |
0.020 |
| 419 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.388 |
-0.044 |
| 420 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.388 |
0.035 |
| 421 |
261203_at |
AT1G12845
|
unknown |
-0.388 |
-0.181 |
| 422 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.386 |
0.065 |
| 423 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.384 |
-0.034 |
| 424 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.384 |
0.107 |
| 425 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.384 |
-0.087 |
| 426 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.384 |
-0.111 |
| 427 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.382 |
0.054 |
| 428 |
249726_at |
AT5G35480
|
unknown |
-0.381 |
-0.067 |
| 429 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.381 |
0.127 |
| 430 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
-0.380 |
-0.015 |
| 431 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-0.379 |
-0.037 |
| 432 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.377 |
-0.167 |
| 433 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.374 |
-0.103 |
| 434 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.372 |
-0.310 |
| 435 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.372 |
-0.095 |
| 436 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.369 |
-0.178 |
| 437 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.368 |
-0.216 |
| 438 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.367 |
-0.087 |
| 439 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.367 |
-0.094 |
| 440 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.365 |
-0.299 |
| 441 |
261598_at |
AT1G49750
|
[Leucine-rich repeat (LRR) family protein] |
-0.365 |
-0.016 |
| 442 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
-0.364 |
-0.169 |
| 443 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.363 |
-0.038 |
| 444 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.363 |
-0.060 |
| 445 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.363 |
-0.009 |
| 446 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.362 |
-0.075 |
| 447 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.361 |
0.022 |
| 448 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.360 |
-0.100 |
| 449 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.360 |
-0.105 |
| 450 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.359 |
-0.397 |
| 451 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
-0.359 |
-0.083 |
| 452 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.359 |
-0.180 |
| 453 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
-0.359 |
-0.092 |
| 454 |
257875_at |
AT3G17120
|
unknown |
-0.356 |
-0.264 |
| 455 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.356 |
-0.108 |
| 456 |
258386_at |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.355 |
0.058 |
| 457 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.354 |
-0.161 |
| 458 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.353 |
-0.190 |
| 459 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.348 |
-0.012 |
| 460 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.348 |
-0.009 |
| 461 |
247878_at |
AT5G57760
|
unknown |
-0.348 |
-0.183 |
| 462 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.346 |
-0.067 |
| 463 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.344 |
-0.208 |
| 464 |
253045_at |
AT4G37445
|
unknown |
-0.342 |
-0.031 |
| 465 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.342 |
-0.155 |
| 466 |
253305_at |
AT4G33666
|
unknown |
-0.339 |
0.064 |
| 467 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.338 |
-0.093 |
| 468 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.337 |
-0.028 |
| 469 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.337 |
-0.147 |
| 470 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.337 |
-0.198 |
| 471 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.335 |
-0.326 |
| 472 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.335 |
0.113 |
| 473 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.334 |
-0.162 |
| 474 |
251036_at |
AT5G02160
|
unknown |
-0.334 |
-0.037 |
| 475 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.333 |
-0.158 |
| 476 |
246487_at |
AT5G16030
|
unknown |
-0.332 |
-0.062 |
| 477 |
248861_at |
AT5G46700
|
TET1, TETRASPANIN 1, TRN2, TORNADO 2 |
-0.332 |
-0.181 |
| 478 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.330 |
-0.012 |
| 479 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.330 |
0.072 |
| 480 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.330 |
-0.075 |
| 481 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.329 |
-0.105 |
| 482 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.329 |
-0.093 |
| 483 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.328 |
-0.142 |
| 484 |
260151_at |
AT1G52910
|
unknown |
-0.328 |
-0.060 |
| 485 |
267279_at |
AT2G19460
|
unknown |
-0.328 |
-0.098 |
| 486 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
-0.328 |
-0.137 |
| 487 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.327 |
0.051 |
| 488 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.327 |
-0.018 |
| 489 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
-0.327 |
-0.117 |
| 490 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.327 |
-0.044 |
| 491 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.327 |
-0.082 |
| 492 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.326 |
-0.091 |
| 493 |
261492_at |
AT1G14290
|
SBH2, sphingoid base hydroxylase 2 |
-0.324 |
-0.116 |
| 494 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.324 |
-0.064 |
| 495 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.323 |
-0.282 |
| 496 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.323 |
-0.294 |
| 497 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.323 |
-0.118 |
| 498 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.321 |
-0.204 |
| 499 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.321 |
0.088 |
| 500 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.321 |
-0.083 |
| 501 |
251058_at |
AT5G01790
|
unknown |
-0.321 |
-0.191 |
| 502 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.319 |
0.090 |
| 503 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.319 |
-0.117 |
| 504 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.319 |
-0.106 |
| 505 |
249709_at |
AT5G35670
|
iqd33, IQ-domain 33 |
-0.318 |
-0.228 |
| 506 |
248646_at |
AT5G49100
|
unknown |
-0.318 |
-0.151 |
| 507 |
249332_at |
AT5G40980
|
unknown |
-0.318 |
-0.150 |
| 508 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
-0.318 |
-0.072 |
| 509 |
250366_at |
AT5G11420
|
unknown |
-0.317 |
-0.094 |
| 510 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.316 |
-0.100 |
| 511 |
257769_at |
AT3G23050
|
AXR2, AUXIN RESISTANT 2, IAA7, indole-3-acetic acid 7 |
-0.316 |
-0.055 |
| 512 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.315 |
-0.206 |
| 513 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.315 |
0.010 |
| 514 |
258535_at |
AT3G06750
|
[hydroxyproline-rich glycoprotein family protein] |
-0.315 |
-0.001 |
| 515 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.315 |
-0.021 |
| 516 |
249986_at |
AT5G18460
|
unknown |
-0.315 |
-0.006 |
| 517 |
261684_at |
AT1G47400
|
unknown |
-0.314 |
0.271 |
| 518 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.314 |
-0.039 |
| 519 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
-0.312 |
0.012 |
| 520 |
255824_at |
AT2G40530
|
unknown |
-0.311 |
-0.167 |
| 521 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.311 |
-0.145 |
| 522 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.310 |
0.071 |
| 523 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.310 |
-0.151 |
| 524 |
251788_at |
AT3G55420
|
unknown |
-0.310 |
-0.228 |
| 525 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.310 |
-0.101 |
| 526 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.309 |
-0.247 |
| 527 |
248139_at |
AT5G54970
|
unknown |
-0.308 |
-0.204 |
| 528 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.308 |
-0.067 |
| 529 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.308 |
0.005 |
| 530 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.307 |
-0.139 |
| 531 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.306 |
-0.060 |
| 532 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.306 |
-0.015 |
| 533 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.306 |
-0.160 |
| 534 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
-0.306 |
-0.191 |
| 535 |
259760_at |
AT1G77580
|
unknown |
-0.305 |
-0.046 |
| 536 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.305 |
0.014 |
| 537 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.305 |
0.006 |
| 538 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
-0.304 |
-0.137 |
| 539 |
255285_at |
AT4G04630
|
unknown |
-0.304 |
-0.185 |
| 540 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.302 |
-0.180 |
| 541 |
262286_at |
AT1G68585
|
unknown |
-0.300 |
-0.112 |
| 542 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
-0.298 |
0.021 |
| 543 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.298 |
-0.007 |
| 544 |
258368_at |
AT3G14240
|
[Subtilase family protein] |
-0.297 |
-0.081 |
| 545 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.296 |
-0.034 |
| 546 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.296 |
0.006 |
| 547 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.294 |
-0.066 |
| 548 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.294 |
0.002 |
| 549 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.293 |
-0.119 |
| 550 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.293 |
-0.205 |
| 551 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.292 |
-0.086 |
| 552 |
258468_at |
AT3G06070
|
unknown |
-0.292 |
-0.061 |
| 553 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.292 |
-0.135 |
| 554 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.291 |
-0.171 |
| 555 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.290 |
-0.198 |
| 556 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.290 |
-0.195 |
| 557 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.290 |
-0.116 |
| 558 |
267344_at |
AT2G44230
|
unknown |
-0.290 |
0.077 |
| 559 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.288 |
-0.046 |
| 560 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.288 |
-0.082 |
| 561 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
-0.288 |
-0.073 |
| 562 |
266311_at |
AT2G27130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.287 |
-0.057 |
| 563 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
-0.287 |
-0.067 |
| 564 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.286 |
-0.264 |
| 565 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.285 |
-0.013 |
| 566 |
254815_at |
AT4G12420
|
SKU5 |
-0.285 |
-0.035 |
| 567 |
246505_at |
AT5G16250
|
unknown |
-0.284 |
-0.188 |
| 568 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.284 |
-0.209 |
| 569 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.283 |
0.012 |
| 570 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.283 |
-0.180 |
| 571 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.282 |
-0.042 |
| 572 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.279 |
-0.135 |
| 573 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.279 |
-0.025 |
| 574 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.278 |
-0.016 |
| 575 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.277 |
-0.187 |
| 576 |
245112_at |
AT2G41540
|
GPDHC1 |
-0.277 |
-0.069 |
| 577 |
251598_at |
AT3G57600
|
[Integrase-type DNA-binding superfamily protein] |
-0.277 |
-0.121 |
| 578 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.277 |
-0.144 |
| 579 |
262884_at |
AT1G64720
|
CP5 |
-0.275 |
0.036 |
| 580 |
258225_at |
AT3G15630
|
unknown |
-0.275 |
-0.014 |
| 581 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.274 |
-0.013 |
| 582 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.274 |
0.006 |
| 583 |
247881_at |
AT5G57700
|
[BNR/Asp-box repeat family protein] |
-0.273 |
-0.187 |
| 584 |
265824_at |
AT2G35650
|
ATCSLA07, cellulose synthase like, ATCSLA7, CSLA07, CSLA07, cellulose synthase like, CSLA7, CELLULOSE SYNTHASE LIKE A7 |
-0.273 |
0.075 |
| 585 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
-0.273 |
-0.022 |
| 586 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.273 |
-0.231 |
| 587 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.272 |
-0.186 |
| 588 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.272 |
-0.372 |
| 589 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
-0.271 |
0.099 |
| 590 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.271 |
-0.343 |
| 591 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
-0.271 |
-0.021 |
| 592 |
262236_at |
AT1G48330
|
unknown |
-0.270 |
-0.266 |
| 593 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.270 |
0.045 |
| 594 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.270 |
0.072 |
| 595 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.270 |
0.103 |
| 596 |
250696_at |
AT5G06790
|
unknown |
-0.269 |
-0.007 |
| 597 |
252852_at |
AT4G39900
|
unknown |
-0.268 |
-0.041 |
| 598 |
252118_at |
AT3G51400
|
unknown |
-0.267 |
-0.080 |
| 599 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.267 |
-0.120 |
| 600 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.267 |
-0.017 |