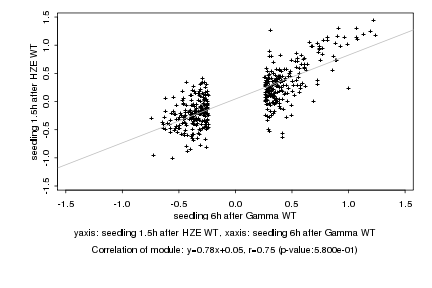
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling 6h after Gamma WT |
Log2 signal ratio
seedling 1.5h after HZE WT |
| 1 |
248086_at |
AT5G55490
|
ATGEX1, GAMETE EXPRESSED PROTEIN 1, GEX1, gamete expressed protein 1 |
1.236 |
1.184 |
| 2 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
1.219 |
1.456 |
| 3 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
1.189 |
1.245 |
| 4 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
1.130 |
1.192 |
| 5 |
259409_at |
AT1G13330
|
AHP2, Arabidopsis Hop2 homolog |
1.072 |
1.111 |
| 6 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
1.064 |
1.305 |
| 7 |
254055_at |
AT4G25330
|
unknown |
1.062 |
1.147 |
| 8 |
252330_at |
AT3G48770
|
[DNA binding] |
0.998 |
0.245 |
| 9 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.986 |
1.018 |
| 10 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.963 |
1.143 |
| 11 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.921 |
0.983 |
| 12 |
252539_at |
AT3G45730
|
unknown |
0.911 |
1.311 |
| 13 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.894 |
1.163 |
| 14 |
254287_at |
AT4G22960
|
unknown |
0.887 |
0.741 |
| 15 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.880 |
1.039 |
| 16 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.880 |
0.734 |
| 17 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.865 |
0.800 |
| 18 |
263515_at |
AT2G21640
|
unknown |
0.850 |
0.556 |
| 19 |
256442_at |
AT3G10930
|
unknown |
0.809 |
1.154 |
| 20 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
0.808 |
1.101 |
| 21 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.773 |
1.099 |
| 22 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.768 |
0.838 |
| 23 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.764 |
0.948 |
| 24 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.748 |
0.736 |
| 25 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.735 |
0.942 |
| 26 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.735 |
0.933 |
| 27 |
250372_at |
AT5G11460
|
unknown |
0.735 |
0.994 |
| 28 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.734 |
0.881 |
| 29 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.725 |
0.317 |
| 30 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.723 |
0.388 |
| 31 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.720 |
1.041 |
| 32 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.690 |
0.011 |
| 33 |
253733_at |
AT4G29170
|
ATMND1 |
0.681 |
0.989 |
| 34 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.666 |
0.977 |
| 35 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.648 |
1.056 |
| 36 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.636 |
0.665 |
| 37 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.620 |
0.563 |
| 38 |
253044_at |
AT4G37290
|
unknown |
0.615 |
0.280 |
| 39 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
0.609 |
0.613 |
| 40 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.607 |
0.675 |
| 41 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
0.605 |
0.777 |
| 42 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.602 |
0.594 |
| 43 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.599 |
0.759 |
| 44 |
259607_at |
AT1G27940
|
ABCB13, ATP-binding cassette B13, PGP13, P-glycoprotein 13 |
0.591 |
0.323 |
| 45 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
0.583 |
0.672 |
| 46 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.578 |
0.828 |
| 47 |
251354_at |
AT3G61090
|
[Putative endonuclease or glycosyl hydrolase] |
0.575 |
0.295 |
| 48 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.569 |
0.238 |
| 49 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.563 |
0.506 |
| 50 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.562 |
0.598 |
| 51 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.558 |
0.164 |
| 52 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.551 |
0.436 |
| 53 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.546 |
0.728 |
| 54 |
253157_at |
AT4G35740
|
ATRECQ3, A. THALIANA RECQ HELICASE 3, RecQl3 |
0.543 |
0.779 |
| 55 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.540 |
0.287 |
| 56 |
247081_at |
AT5G66130
|
ATRAD17, RADIATION SENSITIVE 17, RAD17, RADIATION SENSITIVE 17 |
0.539 |
0.856 |
| 57 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.534 |
0.383 |
| 58 |
248738_at |
AT5G48020
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.532 |
0.618 |
| 59 |
265449_at |
AT2G46610
|
At-RS31a, arginine/serine-rich splicing factor 31a, RS31a, arginine/serine-rich splicing factor 31a |
0.524 |
0.712 |
| 60 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.514 |
0.119 |
| 61 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.505 |
0.292 |
| 62 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.500 |
0.359 |
| 63 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.497 |
0.357 |
| 64 |
252898_at |
AT4G39500
|
CYP96A11, cytochrome P450, family 96, subfamily A, polypeptide 11 |
0.497 |
0.191 |
| 65 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.495 |
-0.058 |
| 66 |
260223_at |
AT1G74390
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.493 |
0.743 |
| 67 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.488 |
-0.241 |
| 68 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.481 |
0.461 |
| 69 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.479 |
0.543 |
| 70 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.471 |
0.508 |
| 71 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.464 |
0.234 |
| 72 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.461 |
0.450 |
| 73 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
0.454 |
0.149 |
| 74 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.454 |
0.003 |
| 75 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.451 |
-0.267 |
| 76 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.447 |
0.571 |
| 77 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.447 |
0.293 |
| 78 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.437 |
0.147 |
| 79 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.437 |
0.417 |
| 80 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.433 |
0.387 |
| 81 |
265212_at |
AT1G05030
|
[Major facilitator superfamily protein] |
0.430 |
0.136 |
| 82 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.430 |
0.579 |
| 83 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.421 |
-0.126 |
| 84 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.419 |
0.299 |
| 85 |
263476_at |
AT2G31870
|
PARG1, poly(ADP-ribose) glycohydrolase 1, TEJ, Sanskrit for 'bright' |
0.413 |
0.458 |
| 86 |
256188_at |
AT1G30160
|
unknown |
0.412 |
0.367 |
| 87 |
252661_at |
AT3G44450
|
unknown |
0.412 |
-0.632 |
| 88 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
0.411 |
0.192 |
| 89 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.409 |
-0.553 |
| 90 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.408 |
0.044 |
| 91 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
0.404 |
0.376 |
| 92 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.401 |
0.545 |
| 93 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.401 |
0.196 |
| 94 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
0.400 |
0.250 |
| 95 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.397 |
0.347 |
| 96 |
250136_at |
AT5G15380
|
DRM1, domains rearranged methylase 1 |
0.396 |
0.448 |
| 97 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.393 |
-0.115 |
| 98 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.391 |
0.830 |
| 99 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.387 |
0.586 |
| 100 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.387 |
-0.072 |
| 101 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.385 |
0.418 |
| 102 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.384 |
-0.018 |
| 103 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.382 |
0.256 |
| 104 |
266595_at |
AT2G46180
|
GC4, golgin candidate 4 |
0.382 |
0.562 |
| 105 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.382 |
0.178 |
| 106 |
245041_at |
AT2G26530
|
AR781 |
0.382 |
0.429 |
| 107 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
0.382 |
0.044 |
| 108 |
267623_at |
AT2G39650
|
unknown |
0.382 |
0.456 |
| 109 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.376 |
0.448 |
| 110 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
0.374 |
0.267 |
| 111 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.371 |
0.163 |
| 112 |
266488_at |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
0.368 |
0.363 |
| 113 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.367 |
0.009 |
| 114 |
251076_at |
AT5G01970
|
unknown |
0.366 |
0.297 |
| 115 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.366 |
0.393 |
| 116 |
266707_at |
AT2G03310
|
unknown |
0.364 |
0.282 |
| 117 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.363 |
0.055 |
| 118 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.361 |
0.409 |
| 119 |
264116_at |
AT2G31320
|
PARP1, POLY(ADP-RIBOSE) POLYMERASE 1 |
0.360 |
0.580 |
| 120 |
262842_at |
AT1G14720
|
ATXTH28, EXGT-A2, ENDOXYLOGLUCAN TRANSFERASE A2, XTH28, xyloglucan endotransglucosylase/hydrolase 28, XTR2, xyloglucan endotransglycosylase related 2 |
0.357 |
0.270 |
| 121 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.357 |
0.344 |
| 122 |
261318_at |
AT1G53035
|
unknown |
0.354 |
-0.021 |
| 123 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
0.353 |
0.172 |
| 124 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.352 |
0.332 |
| 125 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.349 |
0.170 |
| 126 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.349 |
0.477 |
| 127 |
262211_at |
AT1G74930
|
ORA47 |
0.346 |
0.374 |
| 128 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.346 |
-0.002 |
| 129 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.346 |
0.024 |
| 130 |
258045_at |
AT3G21280
|
UBP7, ubiquitin-specific protease 7 |
0.345 |
0.383 |
| 131 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.345 |
-0.169 |
| 132 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.343 |
0.064 |
| 133 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
0.341 |
-0.041 |
| 134 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.340 |
0.160 |
| 135 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.338 |
0.106 |
| 136 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
0.338 |
0.160 |
| 137 |
249355_at |
AT5G40500
|
unknown |
0.336 |
0.230 |
| 138 |
257086_at |
AT3G20490
|
unknown protein; Has 754 Blast hits to 165 proteins in 64 species: Archae - 0; Bacteria - 48; Metazoa - 26; Fungi - 25; Plants - 36; Viruses - 0; Other Eukaryotes - 619 (source: NCBI BLink). |
0.335 |
0.504 |
| 139 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.333 |
0.303 |
| 140 |
264694_at |
AT1G70250
|
[receptor serine/threonine kinase, putative] |
0.331 |
0.122 |
| 141 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.331 |
0.077 |
| 142 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.331 |
0.120 |
| 143 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
0.328 |
-0.009 |
| 144 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
0.328 |
0.698 |
| 145 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.328 |
-0.205 |
| 146 |
263383_at |
AT2G40120
|
[Protein kinase superfamily protein] |
0.324 |
-0.008 |
| 147 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.323 |
0.227 |
| 148 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
0.321 |
0.356 |
| 149 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.320 |
0.292 |
| 150 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.319 |
0.094 |
| 151 |
251058_at |
AT5G01790
|
unknown |
0.318 |
-0.234 |
| 152 |
255223_at |
AT4G05370
|
[BCS1 AAA-type ATPase] |
0.315 |
0.814 |
| 153 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.314 |
0.137 |
| 154 |
267410_at |
AT2G34920
|
EDA18, embryo sac development arrest 18 |
0.313 |
0.550 |
| 155 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
0.313 |
0.188 |
| 156 |
266659_at |
AT2G25850
|
PAPS2, poly(A) polymerase 2 |
0.313 |
0.418 |
| 157 |
247155_at |
AT5G65750
|
[2-oxoglutarate dehydrogenase, E1 component] |
0.310 |
0.247 |
| 158 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.308 |
-0.175 |
| 159 |
261243_at |
AT1G20180
|
unknown |
0.307 |
1.278 |
| 160 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
0.306 |
0.097 |
| 161 |
246997_at |
AT5G67390
|
unknown |
0.304 |
-0.053 |
| 162 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.304 |
0.111 |
| 163 |
251372_at |
AT3G60520
|
unknown |
0.302 |
0.139 |
| 164 |
253830_at |
AT4G27652
|
unknown |
0.301 |
0.810 |
| 165 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.301 |
0.159 |
| 166 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
0.300 |
-0.270 |
| 167 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.300 |
0.487 |
| 168 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.300 |
0.250 |
| 169 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.299 |
-0.032 |
| 170 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.298 |
0.213 |
| 171 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.297 |
0.274 |
| 172 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.296 |
-0.516 |
| 173 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.295 |
0.904 |
| 174 |
246558_at |
AT5G15540
|
ATSCC2, ARABIDOPSIS THALIANA SISTER-CHROMATID COHESION 2, EMB2773, EMBRYO DEFECTIVE 2773, SCC2, SISTER-CHROMATID COHESION 2 |
0.294 |
0.424 |
| 175 |
247080_at |
AT5G66140
|
PAD2, proteasome alpha subunit D2 |
0.294 |
0.295 |
| 176 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
0.292 |
0.470 |
| 177 |
251755_at |
AT3G55790
|
unknown |
0.292 |
-0.486 |
| 178 |
254140_at |
AT4G24610
|
unknown |
0.292 |
0.417 |
| 179 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.290 |
0.178 |
| 180 |
251455_at |
AT3G60100
|
CSY5, citrate synthase 5 |
0.290 |
0.101 |
| 181 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.289 |
0.217 |
| 182 |
248426_at |
AT5G51740
|
[Peptidase family M48 family protein] |
0.289 |
0.404 |
| 183 |
262609_at |
AT1G13930
|
unknown |
0.287 |
0.198 |
| 184 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.285 |
0.369 |
| 185 |
260937_at |
AT1G45160
|
[Protein kinase superfamily protein] |
0.284 |
0.201 |
| 186 |
264838_at |
AT1G03430
|
AHP5, histidine-containing phosphotransfer factor 5 |
0.284 |
0.426 |
| 187 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.283 |
0.106 |
| 188 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.283 |
-0.253 |
| 189 |
248820_at |
AT5G47060
|
unknown |
0.283 |
-0.329 |
| 190 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.283 |
0.355 |
| 191 |
260051_at |
AT1G78210
|
[alpha/beta-Hydrolases superfamily protein] |
0.282 |
-0.040 |
| 192 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.282 |
-0.140 |
| 193 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
0.279 |
0.231 |
| 194 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
0.279 |
0.187 |
| 195 |
266063_at |
AT2G18760
|
CHR8, chromatin remodeling 8 |
0.278 |
0.543 |
| 196 |
264674_at |
AT1G09815
|
POLD4, polymerase delta 4 |
0.278 |
0.321 |
| 197 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.277 |
0.033 |
| 198 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.274 |
0.595 |
| 199 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.274 |
-0.041 |
| 200 |
250700_at |
AT5G06830
|
unknown |
0.273 |
0.384 |
| 201 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
0.273 |
0.224 |
| 202 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.272 |
0.080 |
| 203 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.271 |
0.391 |
| 204 |
261017_at |
AT1G26570
|
ATUGD1, UDP-GLUCOSE DEHYDROGENASE 1, UGD1, UDP-glucose dehydrogenase 1 |
0.269 |
0.110 |
| 205 |
257611_at |
AT3G26580
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.269 |
0.333 |
| 206 |
250417_at |
AT5G11230
|
[Nucleotide-sugar transporter family protein] |
0.268 |
0.051 |
| 207 |
258921_at |
AT3G10500
|
ANAC053, NAC domain containing protein 53, NAC053, NAC domain containing protein 53, NTL4, NAC with transmembrane motif 1-like 4 |
0.267 |
0.387 |
| 208 |
259436_at |
AT1G01500
|
[Erythronate-4-phosphate dehydrogenase family protein] |
0.265 |
0.267 |
| 209 |
257596_at |
AT3G24760
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.264 |
0.298 |
| 210 |
258091_at |
AT3G14560
|
unknown |
0.264 |
0.436 |
| 211 |
259514_at |
AT1G12480
|
CDI3, CARBON DIOXIDE INSENSITIVE 3, OZS1, OZONE-SENSITIVE 1, RCD3, RADICAL-INDUCED CELL DEATH 3, SLAC1, SLOW ANION CHANNEL-ASSOCIATED 1 |
0.264 |
0.442 |
| 212 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.263 |
0.318 |
| 213 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.263 |
-0.247 |
| 214 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.262 |
0.166 |
| 215 |
260772_at |
AT1G49050
|
[Eukaryotic aspartyl protease family protein] |
0.261 |
0.346 |
| 216 |
257723_at |
AT3G18500
|
[DNAse I-like superfamily protein] |
0.261 |
0.024 |
| 217 |
263865_at |
AT2G36910
|
ABCB1, ATP-binding cassette B1, ATPGP1, ARABIDOPSIS THALIANA P GLYCOPROTEIN1, PGP1, P-GLYCOPROTEIN 1 |
0.259 |
0.132 |
| 218 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.259 |
0.080 |
| 219 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.257 |
0.003 |
| 220 |
246450_at |
AT5G16820
|
ATHSF3, ARABIDOPSIS HEAT SHOCK FACTOR 3, ATHSFA1B, ARABIDOPSIS THALIANA CLASS A HEAT SHOCK FACTOR 1B, HSF3, heat shock factor 3, HSFA1B, CLASS A HEAT SHOCK FACTOR 1B |
0.256 |
0.434 |
| 221 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.747 |
-0.292 |
| 222 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.729 |
-0.956 |
| 223 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.647 |
-0.357 |
| 224 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.644 |
-0.397 |
| 225 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.644 |
-0.472 |
| 226 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.629 |
-0.283 |
| 227 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.625 |
-0.175 |
| 228 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.624 |
-0.483 |
| 229 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.619 |
0.063 |
| 230 |
258480_at |
AT3G02640
|
unknown |
-0.601 |
-0.385 |
| 231 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.589 |
-0.404 |
| 232 |
248057_at |
AT5G55520
|
unknown |
-0.582 |
-0.327 |
| 233 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.577 |
-0.465 |
| 234 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.575 |
-0.543 |
| 235 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.568 |
-0.208 |
| 236 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.560 |
-1.008 |
| 237 |
261567_at |
AT1G33055
|
unknown |
-0.553 |
0.075 |
| 238 |
246505_at |
AT5G16250
|
unknown |
-0.549 |
-0.172 |
| 239 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.540 |
-0.500 |
| 240 |
246415_at |
AT5G17160
|
unknown |
-0.539 |
-0.224 |
| 241 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.538 |
-0.218 |
| 242 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.534 |
-0.464 |
| 243 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.529 |
-0.302 |
| 244 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.528 |
-0.431 |
| 245 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.524 |
-0.070 |
| 246 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.522 |
-0.438 |
| 247 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.519 |
-0.550 |
| 248 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.507 |
-0.462 |
| 249 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.504 |
-0.306 |
| 250 |
255460_at |
AT4G02800
|
unknown |
-0.503 |
-0.421 |
| 251 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.502 |
-0.269 |
| 252 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.500 |
-0.467 |
| 253 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.489 |
-0.200 |
| 254 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.483 |
-0.243 |
| 255 |
264636_at |
AT1G65490
|
unknown |
-0.477 |
-0.577 |
| 256 |
257794_at |
AT3G27050
|
unknown |
-0.476 |
0.028 |
| 257 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-0.470 |
0.188 |
| 258 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.469 |
-0.382 |
| 259 |
252089_at |
AT3G52110
|
unknown |
-0.468 |
-0.268 |
| 260 |
262081_at |
AT1G59540
|
ZCF125 |
-0.467 |
-0.317 |
| 261 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.462 |
-0.025 |
| 262 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.461 |
-0.038 |
| 263 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.461 |
0.071 |
| 264 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.459 |
-0.362 |
| 265 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.456 |
-0.253 |
| 266 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.455 |
-0.589 |
| 267 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.454 |
-0.553 |
| 268 |
267636_at |
AT2G42110
|
unknown |
-0.448 |
-0.374 |
| 269 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.448 |
-0.191 |
| 270 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.448 |
-0.243 |
| 271 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
-0.447 |
-0.153 |
| 272 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.447 |
-0.508 |
| 273 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.446 |
-0.292 |
| 274 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.439 |
-0.061 |
| 275 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.438 |
-0.786 |
| 276 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
-0.437 |
-0.421 |
| 277 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.435 |
0.355 |
| 278 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.431 |
-0.880 |
| 279 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.428 |
-0.586 |
| 280 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.422 |
-0.191 |
| 281 |
245953_at |
AT5G28520
|
[Mannose-binding lectin superfamily protein] |
-0.413 |
-0.162 |
| 282 |
253978_at |
AT4G26660
|
unknown |
-0.407 |
-0.306 |
| 283 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.405 |
-0.838 |
| 284 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.403 |
-0.149 |
| 285 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.402 |
-0.191 |
| 286 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.402 |
0.116 |
| 287 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.399 |
-0.224 |
| 288 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.396 |
-0.237 |
| 289 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.394 |
-0.621 |
| 290 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.393 |
-0.058 |
| 291 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.393 |
0.021 |
| 292 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.393 |
0.121 |
| 293 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.391 |
-0.319 |
| 294 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.390 |
-0.297 |
| 295 |
257134_at |
AT3G12870
|
unknown |
-0.389 |
-0.270 |
| 296 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
-0.388 |
-0.485 |
| 297 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.387 |
-0.639 |
| 298 |
251010_at |
AT5G02550
|
unknown |
-0.387 |
-0.262 |
| 299 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.386 |
0.205 |
| 300 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.386 |
-0.575 |
| 301 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.383 |
-0.380 |
| 302 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.381 |
-0.200 |
| 303 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.381 |
-0.058 |
| 304 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
-0.379 |
-0.040 |
| 305 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.379 |
-0.399 |
| 306 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.378 |
0.280 |
| 307 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.377 |
-0.677 |
| 308 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.377 |
-0.294 |
| 309 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.376 |
-0.297 |
| 310 |
256603_at |
AT3G28270
|
unknown |
-0.376 |
-0.584 |
| 311 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.375 |
-0.340 |
| 312 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.373 |
-0.250 |
| 313 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.369 |
-0.172 |
| 314 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.368 |
-0.162 |
| 315 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.368 |
-0.081 |
| 316 |
258867_at |
AT3G03130
|
unknown |
-0.367 |
-0.304 |
| 317 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.366 |
0.183 |
| 318 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
-0.365 |
-0.403 |
| 319 |
253804_at |
AT4G28230
|
unknown |
-0.361 |
0.147 |
| 320 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.361 |
-0.202 |
| 321 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.359 |
-0.228 |
| 322 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
-0.358 |
-0.156 |
| 323 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.356 |
-0.319 |
| 324 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
-0.355 |
-0.195 |
| 325 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.353 |
-0.042 |
| 326 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.352 |
-0.162 |
| 327 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.350 |
-0.109 |
| 328 |
254132_at |
AT4G24660
|
ATHB22, HOMEOBOX PROTEIN 22, HB22, homeobox protein 22, MEE68, MATERNAL EFFECT EMBRYO ARREST 68, ZHD2, ZINC FINGER HOMEODOMAIN 2 |
-0.349 |
-0.199 |
| 329 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.349 |
-0.321 |
| 330 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.348 |
-0.287 |
| 331 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.347 |
-0.410 |
| 332 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.346 |
-0.303 |
| 333 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.339 |
-0.384 |
| 334 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.337 |
-0.639 |
| 335 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
-0.335 |
0.035 |
| 336 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.335 |
-0.293 |
| 337 |
248691_at |
AT5G48310
|
unknown |
-0.334 |
-0.173 |
| 338 |
245713_at |
AT5G04370
|
NAMT1 |
-0.333 |
-0.202 |
| 339 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.330 |
-0.553 |
| 340 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.330 |
-0.308 |
| 341 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.329 |
0.157 |
| 342 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.326 |
-0.581 |
| 343 |
258397_at |
AT3G15357
|
unknown |
-0.324 |
-0.327 |
| 344 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.322 |
-0.496 |
| 345 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-0.321 |
0.193 |
| 346 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.321 |
0.113 |
| 347 |
253919_at |
AT4G27350
|
unknown |
-0.320 |
-0.193 |
| 348 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
-0.319 |
-0.217 |
| 349 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.317 |
-0.230 |
| 350 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.316 |
-0.497 |
| 351 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.316 |
-0.176 |
| 352 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.315 |
0.228 |
| 353 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.315 |
-0.189 |
| 354 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.315 |
0.098 |
| 355 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.314 |
0.004 |
| 356 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.313 |
-0.094 |
| 357 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
-0.313 |
0.308 |
| 358 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
-0.312 |
-0.084 |
| 359 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.310 |
-0.773 |
| 360 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.309 |
-0.169 |
| 361 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.306 |
-0.082 |
| 362 |
256666_at |
AT3G20670
|
HTA13, histone H2A 13 |
-0.306 |
-0.142 |
| 363 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.306 |
-0.400 |
| 364 |
250386_at |
AT5G11510
|
AtMYB3R4, myb domain protein 3R4, MYB3R-4, myb domain protein 3r-4 |
-0.305 |
-0.178 |
| 365 |
245353_at |
AT4G16000
|
unknown |
-0.304 |
0.320 |
| 366 |
247039_at |
AT5G67270
|
ATEB1C, MICROTUBULE END BINDING PROTEIN 1, ATEB1H1, ATEB1-HOMOLOG1, EB1C, end binding protein 1C |
-0.304 |
-0.094 |
| 367 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.303 |
-0.195 |
| 368 |
251363_at |
AT3G61250
|
AtMYB17, myb domain protein 17, LMI2, LATE MERISTEM IDENTITY2, MYB17, myb domain protein 17 |
-0.302 |
-0.121 |
| 369 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
-0.300 |
-0.471 |
| 370 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.300 |
-0.342 |
| 371 |
264238_at |
AT1G54740
|
unknown |
-0.298 |
0.348 |
| 372 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.297 |
0.159 |
| 373 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.297 |
-0.353 |
| 374 |
261658_at |
AT1G50040
|
unknown |
-0.296 |
0.423 |
| 375 |
267613_at |
AT2G26700
|
PID2, PINOID2 |
-0.294 |
-0.131 |
| 376 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.291 |
0.086 |
| 377 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.288 |
-0.022 |
| 378 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.286 |
-0.209 |
| 379 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.286 |
0.139 |
| 380 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.285 |
-0.143 |
| 381 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.283 |
0.073 |
| 382 |
247563_at |
AT5G61130
|
AtPDCB1, PDCB1, plasmodesmata callose-binding protein 1 |
-0.283 |
-0.246 |
| 383 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.282 |
-0.342 |
| 384 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
-0.282 |
-0.446 |
| 385 |
259316_at |
AT3G01175
|
unknown |
-0.281 |
0.348 |
| 386 |
246783_at |
AT5G27360
|
SFP2 |
-0.280 |
-0.324 |
| 387 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
-0.280 |
-0.063 |
| 388 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.278 |
-0.197 |
| 389 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.278 |
0.045 |
| 390 |
258326_at |
AT3G22760
|
SOL1 |
-0.277 |
-0.187 |
| 391 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.277 |
-0.334 |
| 392 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
-0.276 |
-0.169 |
| 393 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.276 |
0.047 |
| 394 |
249189_at |
AT5G42780
|
AtHB27, homeobox protein 27, HB27, homeobox protein 27, ZHD13, ZINC FINGER HOMEODOMAIN 13 |
-0.275 |
0.019 |
| 395 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.274 |
-0.052 |
| 396 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.273 |
-0.392 |
| 397 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.273 |
0.263 |
| 398 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.273 |
-0.115 |
| 399 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.273 |
-0.257 |
| 400 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.268 |
-0.442 |
| 401 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.268 |
0.053 |
| 402 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.268 |
-0.251 |
| 403 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.267 |
0.114 |
| 404 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.267 |
-0.670 |
| 405 |
262777_at |
AT1G13030
|
Atcoilin, COILIN, coilin |
-0.266 |
-0.152 |
| 406 |
253817_at |
AT4G28310
|
unknown |
-0.265 |
-0.153 |
| 407 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.265 |
0.163 |
| 408 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.264 |
-0.493 |
| 409 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.264 |
-0.454 |
| 410 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.262 |
-0.809 |
| 411 |
254193_at |
AT4G23870
|
unknown |
-0.261 |
0.303 |
| 412 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.261 |
-0.089 |
| 413 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
-0.260 |
-0.121 |
| 414 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.260 |
-0.203 |
| 415 |
254561_at |
AT4G19160
|
unknown |
-0.258 |
0.042 |
| 416 |
247329_at |
AT5G64150
|
[RNA methyltransferase family protein] |
-0.257 |
-0.065 |
| 417 |
261296_at |
AT1G48460
|
unknown |
-0.257 |
-0.391 |
| 418 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-0.256 |
-0.054 |
| 419 |
253317_at |
AT4G33960
|
unknown |
-0.256 |
-0.370 |
| 420 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.255 |
0.133 |
| 421 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
-0.254 |
-0.122 |
| 422 |
256861_at |
AT3G23920
|
AtBAM1, BAM1, beta-amylase 1, BMY7, BETA-AMYLASE 7, TR-BAMY |
-0.254 |
0.018 |
| 423 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.254 |
-0.479 |
| 424 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.253 |
-0.100 |
| 425 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.253 |
-0.328 |
| 426 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
-0.253 |
-0.041 |
| 427 |
247944_at |
AT5G57100
|
[Nucleotide/sugar transporter family protein] |
-0.252 |
-0.028 |
| 428 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.252 |
0.136 |
| 429 |
254400_at |
AT4G21270
|
ATK1, kinesin 1, KATA, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A, KATAP, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A PROTEIN |
-0.251 |
-0.206 |
| 430 |
264176_at |
AT1G02110
|
unknown |
-0.251 |
0.179 |
| 431 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.251 |
-0.316 |
| 432 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.249 |
-0.069 |
| 433 |
247599_at |
AT5G60880
|
BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE |
-0.248 |
-0.141 |
| 434 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.248 |
-0.210 |
| 435 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.248 |
-0.215 |
| 436 |
246926_at |
AT5G25240
|
unknown |
-0.247 |
0.107 |
| 437 |
266264_at |
AT2G27775
|
unknown |
-0.247 |
-0.233 |
| 438 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.245 |
-0.460 |
| 439 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.245 |
-0.404 |