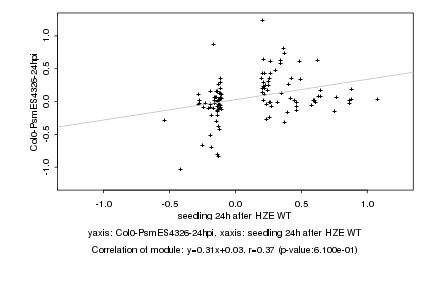
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling 24h after HZE WT |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
1.074 |
0.033 |
| 2 |
252539_at |
AT3G45730
|
unknown |
0.877 |
0.194 |
| 3 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.877 |
0.034 |
| 4 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.862 |
0.027 |
| 5 |
248086_at |
AT5G55490
|
ATGEX1, GAMETE EXPRESSED PROTEIN 1, GEX1, gamete expressed protein 1 |
0.859 |
-0.030 |
| 6 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.764 |
0.076 |
| 7 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.747 |
-0.140 |
| 8 |
254055_at |
AT4G25330
|
unknown |
0.645 |
0.176 |
| 9 |
254287_at |
AT4G22960
|
unknown |
0.644 |
0.085 |
| 10 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.626 |
0.090 |
| 11 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.619 |
0.627 |
| 12 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.602 |
-0.014 |
| 13 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.599 |
0.028 |
| 14 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.590 |
0.027 |
| 15 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
0.573 |
-0.051 |
| 16 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.488 |
0.337 |
| 17 |
253044_at |
AT4G37290
|
unknown |
0.485 |
0.618 |
| 18 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.463 |
-0.001 |
| 19 |
254565_at |
AT4G19130
|
[Replication factor-A protein 1-related] |
0.459 |
-0.064 |
| 20 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
0.456 |
-0.134 |
| 21 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.443 |
0.021 |
| 22 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.423 |
0.355 |
| 23 |
247081_at |
AT5G66130
|
ATRAD17, RADIATION SENSITIVE 17, RAD17, RADIATION SENSITIVE 17 |
0.417 |
0.048 |
| 24 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.398 |
0.274 |
| 25 |
250372_at |
AT5G11460
|
unknown |
0.392 |
-0.165 |
| 26 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
0.370 |
-0.311 |
| 27 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.370 |
0.737 |
| 28 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.362 |
0.818 |
| 29 |
252330_at |
AT3G48770
|
[DNA binding] |
0.346 |
0.137 |
| 30 |
263515_at |
AT2G21640
|
unknown |
0.340 |
0.587 |
| 31 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.337 |
0.639 |
| 32 |
253157_at |
AT4G35740
|
ATRECQ3, A. THALIANA RECQ HELICASE 3, RecQl3 |
0.314 |
-0.003 |
| 33 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.302 |
0.484 |
| 34 |
265449_at |
AT2G46610
|
At-RS31a, arginine/serine-rich splicing factor 31a, RS31a, arginine/serine-rich splicing factor 31a |
0.267 |
-0.074 |
| 35 |
260477_at |
AT1G11050
|
[Protein kinase superfamily protein] |
0.263 |
0.437 |
| 36 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.262 |
0.610 |
| 37 |
247528_at |
AT5G61460
|
ATRAD18, MIM, hypersensitive to MMS, irradiation and MMC, SMC6B, STRUCTURAL MAINTENANCE OF CHROMOSOMES 6B |
0.259 |
-0.001 |
| 38 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.253 |
0.354 |
| 39 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.252 |
-0.007 |
| 40 |
264116_at |
AT2G31320
|
PARP1, POLY(ADP-RIBOSE) POLYMERASE 1 |
0.251 |
-0.234 |
| 41 |
259792_at |
AT1G29690
|
CAD1, constitutively activated cell death 1 |
0.249 |
0.309 |
| 42 |
248738_at |
AT5G48020
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.245 |
0.250 |
| 43 |
251636_at |
AT3G57530
|
ATCPK32, CDPK32, CPK32, calcium-dependent protein kinase 32 |
0.239 |
0.181 |
| 44 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
0.232 |
-0.264 |
| 45 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.230 |
-0.035 |
| 46 |
254592_at |
AT4G18880
|
AT-HSFA4A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A4A, HSF A4A, heat shock transcription factor A4A |
0.221 |
0.246 |
| 47 |
261063_at |
AT1G07520
|
[GRAS family transcription factor] |
0.218 |
0.439 |
| 48 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.217 |
0.111 |
| 49 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.216 |
0.206 |
| 50 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.210 |
0.024 |
| 51 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.210 |
0.240 |
| 52 |
247080_at |
AT5G66140
|
PAD2, proteasome alpha subunit D2 |
0.210 |
0.292 |
| 53 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.209 |
0.643 |
| 54 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.205 |
0.139 |
| 55 |
247426_at |
AT5G62570
|
CBP60a, calmodulin-binding protein 60a |
0.203 |
0.212 |
| 56 |
245353_at |
AT4G16000
|
unknown |
0.200 |
0.435 |
| 57 |
245209_at |
AT5G12340
|
unknown |
0.198 |
1.249 |
| 58 |
259602_at |
AT1G56520
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.196 |
0.356 |
| 59 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.543 |
-0.284 |
| 60 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.423 |
-1.026 |
| 61 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.284 |
0.120 |
| 62 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.283 |
-0.037 |
| 63 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.280 |
-0.030 |
| 64 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
-0.275 |
0.025 |
| 65 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.256 |
-0.663 |
| 66 |
257134_at |
AT3G12870
|
unknown |
-0.245 |
-0.078 |
| 67 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.233 |
-0.026 |
| 68 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.211 |
-0.096 |
| 69 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.195 |
0.154 |
| 70 |
248710_at |
AT5G48480
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.194 |
-0.033 |
| 71 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.192 |
-0.506 |
| 72 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.191 |
-0.082 |
| 73 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.189 |
-0.689 |
| 74 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.182 |
-0.200 |
| 75 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.169 |
-0.103 |
| 76 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
-0.167 |
0.873 |
| 77 |
256126_at |
AT1G18050
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
-0.164 |
0.027 |
| 78 |
255907_at |
AT1G17920
|
HDG12, homeodomain GLABROUS 12 |
-0.161 |
-0.024 |
| 79 |
263040_at |
AT1G23300
|
[MATE efflux family protein] |
-0.159 |
0.062 |
| 80 |
253661_at |
AT4G30130
|
unknown |
-0.155 |
0.063 |
| 81 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.148 |
0.161 |
| 82 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.147 |
-0.292 |
| 83 |
258587_at |
AT3G04310
|
unknown |
-0.144 |
0.071 |
| 84 |
266427_at |
AT2G07170
|
[ARM repeat superfamily protein] |
-0.143 |
-0.063 |
| 85 |
250758_at |
AT5G06000
|
ATEIF3G2, ARABIDOPSIS THALIANA EUKARYOTIC TRANSLATION INITIATION FACTOR 3G2, EIF3G2, eukaryotic translation initiation factor 3G2 |
-0.141 |
-0.139 |
| 86 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
-0.141 |
0.159 |
| 87 |
250937_at |
AT5G03230
|
unknown |
-0.140 |
-0.204 |
| 88 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.137 |
-0.129 |
| 89 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.137 |
0.002 |
| 90 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.137 |
-0.802 |
| 91 |
248631_at |
AT5G49000
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.136 |
0.267 |
| 92 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.133 |
-0.108 |
| 93 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.133 |
-0.370 |
| 94 |
247244_at |
AT5G64710
|
[Putative endonuclease or glycosyl hydrolase] |
-0.133 |
0.027 |
| 95 |
263787_at |
AT2G46420
|
unknown |
-0.132 |
0.020 |
| 96 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.132 |
-0.043 |
| 97 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.131 |
-0.834 |
| 98 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
-0.125 |
-0.098 |
| 99 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.124 |
-0.412 |
| 100 |
264743_at |
AT1G62090
|
[pseudogene] |
-0.124 |
0.137 |
| 101 |
249571_at |
AT5G37620
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.122 |
0.124 |
| 102 |
256490_at |
AT1G31460
|
unknown |
-0.121 |
-0.033 |
| 103 |
266629_at |
AT2G35590
|
[pseudogene] |
-0.121 |
0.019 |
| 104 |
266340_at |
AT2G01480
|
[O-fucosyltransferase family protein] |
-0.120 |
0.302 |
| 105 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
-0.119 |
0.052 |
| 106 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.119 |
0.358 |
| 107 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.119 |
-0.056 |
| 108 |
261306_at |
AT1G48610
|
[AT hook motif-containing protein] |
-0.118 |
-0.078 |
| 109 |
249169_at |
AT5G42880
|
unknown |
-0.116 |
0.060 |
| 110 |
249673_at |
AT5G35920
|
pseudogene |
-0.115 |
0.068 |
| 111 |
259583_at |
AT1G28070
|
unknown |
-0.114 |
0.134 |
| 112 |
256212_at |
AT1G50970
|
[Membrane trafficking VPS53 family protein] |
-0.114 |
0.200 |
| 113 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
-0.111 |
0.050 |
| 114 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.110 |
0.118 |
| 115 |
253516_at |
AT4G31360
|
[selenium binding] |
-0.109 |
-0.117 |