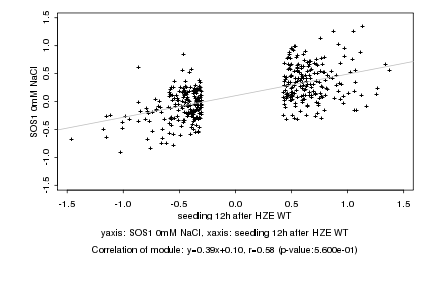
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling 12h after HZE WT |
Log2 signal ratio
SOS1 0mM NaCl |
| 1 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
1.373 |
0.557 |
| 2 |
256442_at |
AT3G10930
|
unknown |
1.331 |
0.665 |
| 3 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
1.258 |
0.249 |
| 4 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
1.253 |
0.128 |
| 5 |
248086_at |
AT5G55490
|
ATGEX1, GAMETE EXPRESSED PROTEIN 1, GEX1, gamete expressed protein 1 |
1.165 |
-0.079 |
| 6 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
1.127 |
1.346 |
| 7 |
253830_at |
AT4G27652
|
unknown |
1.119 |
0.110 |
| 8 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
1.109 |
0.895 |
| 9 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
1.074 |
-0.152 |
| 10 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
1.068 |
0.563 |
| 11 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
1.065 |
0.349 |
| 12 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
1.061 |
-0.154 |
| 13 |
253859_at |
AT4G27657
|
unknown |
1.056 |
0.187 |
| 14 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
1.050 |
1.258 |
| 15 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
1.038 |
0.757 |
| 16 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
1.020 |
0.524 |
| 17 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
1.001 |
0.159 |
| 18 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.968 |
0.820 |
| 19 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.967 |
0.956 |
| 20 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.962 |
-0.034 |
| 21 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.959 |
0.103 |
| 22 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.937 |
0.315 |
| 23 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.933 |
0.044 |
| 24 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.928 |
0.682 |
| 25 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.924 |
0.332 |
| 26 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
0.914 |
0.187 |
| 27 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.912 |
1.027 |
| 28 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.900 |
0.483 |
| 29 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.895 |
0.297 |
| 30 |
254055_at |
AT4G25330
|
unknown |
0.877 |
0.071 |
| 31 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.868 |
1.258 |
| 32 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.862 |
0.425 |
| 33 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.854 |
0.555 |
| 34 |
252539_at |
AT3G45730
|
unknown |
0.837 |
0.440 |
| 35 |
257925_at |
AT3G23170
|
unknown |
0.814 |
0.467 |
| 36 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.813 |
0.233 |
| 37 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.804 |
0.237 |
| 38 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.798 |
0.390 |
| 39 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
0.794 |
0.120 |
| 40 |
253643_at |
AT4G29780
|
unknown |
0.790 |
0.790 |
| 41 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.787 |
-0.151 |
| 42 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.786 |
0.537 |
| 43 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.783 |
0.527 |
| 44 |
262211_at |
AT1G74930
|
ORA47 |
0.776 |
0.668 |
| 45 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.774 |
0.084 |
| 46 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.772 |
0.267 |
| 47 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.765 |
0.284 |
| 48 |
253322_at |
AT4G33980
|
unknown |
0.763 |
0.772 |
| 49 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.763 |
-0.296 |
| 50 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.759 |
0.107 |
| 51 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.758 |
0.542 |
| 52 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.758 |
0.554 |
| 53 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.755 |
0.159 |
| 54 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.752 |
1.138 |
| 55 |
259409_at |
AT1G13330
|
AHP2, Arabidopsis Hop2 homolog |
0.746 |
0.064 |
| 56 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.746 |
0.352 |
| 57 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.735 |
-0.025 |
| 58 |
254287_at |
AT4G22960
|
unknown |
0.732 |
-0.070 |
| 59 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.730 |
0.655 |
| 60 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.728 |
-0.203 |
| 61 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.724 |
0.512 |
| 62 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
0.721 |
0.361 |
| 63 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.719 |
0.754 |
| 64 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.717 |
-0.002 |
| 65 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.716 |
0.673 |
| 66 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.715 |
0.371 |
| 67 |
250098_at |
AT5G17350
|
unknown |
0.712 |
0.133 |
| 68 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.710 |
0.161 |
| 69 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.707 |
-0.252 |
| 70 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.707 |
0.127 |
| 71 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.702 |
0.515 |
| 72 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.698 |
0.696 |
| 73 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.692 |
0.313 |
| 74 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.692 |
0.180 |
| 75 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.675 |
0.119 |
| 76 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.674 |
0.577 |
| 77 |
247177_at |
AT5G65300
|
unknown |
0.670 |
0.256 |
| 78 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.670 |
0.074 |
| 79 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.665 |
0.734 |
| 80 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.665 |
0.425 |
| 81 |
247047_at |
AT5G66650
|
unknown |
0.663 |
0.318 |
| 82 |
245041_at |
AT2G26530
|
AR781 |
0.662 |
0.570 |
| 83 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.660 |
0.720 |
| 84 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.659 |
0.117 |
| 85 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.657 |
0.477 |
| 86 |
252330_at |
AT3G48770
|
[DNA binding] |
0.654 |
0.179 |
| 87 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.652 |
0.679 |
| 88 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.651 |
0.631 |
| 89 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
0.638 |
-0.075 |
| 90 |
267623_at |
AT2G39650
|
unknown |
0.637 |
0.319 |
| 91 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.635 |
0.552 |
| 92 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.635 |
0.707 |
| 93 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
0.634 |
-0.067 |
| 94 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.632 |
0.282 |
| 95 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.631 |
-0.234 |
| 96 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.630 |
0.417 |
| 97 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.627 |
0.364 |
| 98 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.623 |
0.482 |
| 99 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.623 |
0.233 |
| 100 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
0.619 |
0.108 |
| 101 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.617 |
0.903 |
| 102 |
247081_at |
AT5G66130
|
ATRAD17, RADIATION SENSITIVE 17, RAD17, RADIATION SENSITIVE 17 |
0.612 |
0.053 |
| 103 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.608 |
0.745 |
| 104 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.607 |
0.205 |
| 105 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.604 |
0.845 |
| 106 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
0.602 |
0.053 |
| 107 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.600 |
0.299 |
| 108 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.600 |
0.635 |
| 109 |
259850_at |
AT1G72240
|
unknown |
0.600 |
0.122 |
| 110 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.599 |
0.425 |
| 111 |
259607_at |
AT1G27940
|
ABCB13, ATP-binding cassette B13, PGP13, P-glycoprotein 13 |
0.597 |
-0.003 |
| 112 |
253044_at |
AT4G37290
|
unknown |
0.597 |
0.251 |
| 113 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.595 |
0.454 |
| 114 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.595 |
0.346 |
| 115 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.594 |
0.019 |
| 116 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.585 |
0.644 |
| 117 |
253292_at |
AT4G33985
|
unknown |
0.585 |
0.607 |
| 118 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.584 |
0.421 |
| 119 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.583 |
0.489 |
| 120 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.580 |
0.092 |
| 121 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.578 |
0.269 |
| 122 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.574 |
-0.169 |
| 123 |
247800_at |
AT5G58570
|
unknown |
0.573 |
0.382 |
| 124 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.569 |
0.522 |
| 125 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.562 |
0.423 |
| 126 |
267036_at |
AT2G38465
|
unknown |
0.561 |
0.672 |
| 127 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.553 |
0.567 |
| 128 |
263182_at |
AT1G05575
|
unknown |
0.553 |
0.837 |
| 129 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.552 |
0.653 |
| 130 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.552 |
0.089 |
| 131 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.550 |
0.609 |
| 132 |
260688_at |
AT1G17665
|
unknown |
0.549 |
0.549 |
| 133 |
263829_at |
AT2G40435
|
unknown |
0.547 |
-0.315 |
| 134 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.547 |
0.626 |
| 135 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.546 |
0.371 |
| 136 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.542 |
0.796 |
| 137 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.539 |
0.207 |
| 138 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.534 |
0.354 |
| 139 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.534 |
-0.023 |
| 140 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.534 |
-0.083 |
| 141 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.531 |
1.001 |
| 142 |
260656_at |
AT1G19380
|
unknown |
0.527 |
0.415 |
| 143 |
254520_at |
AT4G19960
|
ATKUP9, HAK9, KT9, KUP9, K+ uptake permease 9 |
0.526 |
0.300 |
| 144 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
0.523 |
0.155 |
| 145 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.522 |
0.992 |
| 146 |
250372_at |
AT5G11460
|
unknown |
0.522 |
-0.292 |
| 147 |
265184_at |
AT1G23710
|
unknown |
0.519 |
0.554 |
| 148 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.516 |
0.930 |
| 149 |
260227_at |
AT1G74450
|
unknown |
0.508 |
0.179 |
| 150 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.508 |
0.261 |
| 151 |
248738_at |
AT5G48020
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.507 |
0.118 |
| 152 |
253157_at |
AT4G35740
|
ATRECQ3, A. THALIANA RECQ HELICASE 3, RecQl3 |
0.506 |
0.050 |
| 153 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.502 |
0.103 |
| 154 |
246929_at |
AT5G25210
|
unknown |
0.502 |
0.949 |
| 155 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
0.500 |
-0.237 |
| 156 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.498 |
0.030 |
| 157 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.498 |
0.465 |
| 158 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.497 |
0.138 |
| 159 |
263613_at |
AT2G25250
|
unknown |
0.496 |
0.251 |
| 160 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
0.495 |
0.178 |
| 161 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.495 |
0.674 |
| 162 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.495 |
0.475 |
| 163 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.494 |
0.964 |
| 164 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.490 |
0.786 |
| 165 |
254592_at |
AT4G18880
|
AT-HSFA4A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A4A, HSF A4A, heat shock transcription factor A4A |
0.490 |
0.342 |
| 166 |
247708_at |
AT5G59550
|
unknown |
0.490 |
0.454 |
| 167 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
0.489 |
0.074 |
| 168 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.488 |
0.030 |
| 169 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.484 |
0.656 |
| 170 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.483 |
0.581 |
| 171 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.481 |
0.599 |
| 172 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.479 |
0.881 |
| 173 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.475 |
0.850 |
| 174 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.473 |
0.308 |
| 175 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.472 |
0.355 |
| 176 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.470 |
0.792 |
| 177 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.470 |
0.672 |
| 178 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.469 |
0.111 |
| 179 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
0.468 |
0.582 |
| 180 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
0.459 |
0.153 |
| 181 |
262452_at |
AT1G11210
|
unknown |
0.459 |
0.788 |
| 182 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.456 |
0.105 |
| 183 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.450 |
0.269 |
| 184 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.449 |
0.443 |
| 185 |
260223_at |
AT1G74390
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.447 |
0.126 |
| 186 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
0.447 |
0.062 |
| 187 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.447 |
0.098 |
| 188 |
266488_at |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
0.447 |
-0.308 |
| 189 |
263931_at |
AT2G36220
|
unknown |
0.445 |
0.405 |
| 190 |
265449_at |
AT2G46610
|
At-RS31a, arginine/serine-rich splicing factor 31a, RS31a, arginine/serine-rich splicing factor 31a |
0.441 |
-0.092 |
| 191 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
0.438 |
0.215 |
| 192 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
0.436 |
0.294 |
| 193 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.435 |
0.370 |
| 194 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.432 |
0.685 |
| 195 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.431 |
0.291 |
| 196 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.430 |
0.452 |
| 197 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.430 |
-0.036 |
| 198 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.429 |
0.036 |
| 199 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.428 |
0.163 |
| 200 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
0.427 |
-0.235 |
| 201 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.424 |
0.080 |
| 202 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-1.468 |
-0.674 |
| 203 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-1.183 |
-0.493 |
| 204 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-1.154 |
-0.265 |
| 205 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-1.150 |
-0.633 |
| 206 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-1.117 |
-0.249 |
| 207 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-1.026 |
-0.895 |
| 208 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-1.008 |
-0.475 |
| 209 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-1.007 |
-0.365 |
| 210 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.986 |
-0.256 |
| 211 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.953 |
-0.307 |
| 212 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-0.873 |
-0.012 |
| 213 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.869 |
0.626 |
| 214 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.868 |
-0.350 |
| 215 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.854 |
-0.173 |
| 216 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.802 |
-0.314 |
| 217 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.796 |
-0.116 |
| 218 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.788 |
-0.175 |
| 219 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.788 |
-0.676 |
| 220 |
259373_at |
AT1G69160
|
unknown |
-0.779 |
-0.213 |
| 221 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.773 |
-0.348 |
| 222 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.759 |
-0.825 |
| 223 |
256096_at |
AT1G13650
|
unknown |
-0.743 |
-0.183 |
| 224 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.743 |
-0.520 |
| 225 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.714 |
0.048 |
| 226 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.711 |
-0.147 |
| 227 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.708 |
-0.136 |
| 228 |
247474_at |
AT5G62280
|
unknown |
-0.687 |
-0.081 |
| 229 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.678 |
0.013 |
| 230 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.676 |
-0.090 |
| 231 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.664 |
-0.734 |
| 232 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.662 |
-0.190 |
| 233 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.652 |
-0.648 |
| 234 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.651 |
-0.499 |
| 235 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.641 |
-0.319 |
| 236 |
250327_at |
AT5G12050
|
unknown |
-0.619 |
-0.738 |
| 237 |
249932_at |
AT5G22390
|
unknown |
-0.601 |
-0.103 |
| 238 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.596 |
0.165 |
| 239 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
-0.596 |
-0.386 |
| 240 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.595 |
0.094 |
| 241 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.591 |
-0.558 |
| 242 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.585 |
0.229 |
| 243 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.582 |
-0.302 |
| 244 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.581 |
-0.122 |
| 245 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.579 |
0.135 |
| 246 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.576 |
-0.310 |
| 247 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.575 |
-0.275 |
| 248 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.573 |
-0.415 |
| 249 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.573 |
0.267 |
| 250 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.566 |
0.027 |
| 251 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.563 |
0.108 |
| 252 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.555 |
-0.787 |
| 253 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.554 |
-0.581 |
| 254 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.553 |
0.264 |
| 255 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.552 |
0.063 |
| 256 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.548 |
-0.289 |
| 257 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.546 |
0.376 |
| 258 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.537 |
-0.074 |
| 259 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.536 |
-0.045 |
| 260 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.534 |
0.123 |
| 261 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.524 |
-0.260 |
| 262 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
-0.520 |
0.056 |
| 263 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.517 |
0.021 |
| 264 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.516 |
-0.145 |
| 265 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.513 |
0.026 |
| 266 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.511 |
-0.042 |
| 267 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.509 |
-0.330 |
| 268 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.503 |
0.189 |
| 269 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.493 |
0.262 |
| 270 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.493 |
-0.599 |
| 271 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.489 |
-0.430 |
| 272 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.484 |
-0.076 |
| 273 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.475 |
0.171 |
| 274 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.475 |
0.003 |
| 275 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.474 |
-0.108 |
| 276 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.472 |
0.563 |
| 277 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.470 |
-0.332 |
| 278 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.469 |
0.075 |
| 279 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.469 |
0.851 |
| 280 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.466 |
-0.369 |
| 281 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.465 |
0.249 |
| 282 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.461 |
-0.180 |
| 283 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.459 |
-0.059 |
| 284 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.458 |
0.190 |
| 285 |
255460_at |
AT4G02800
|
unknown |
-0.457 |
0.142 |
| 286 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.456 |
-0.331 |
| 287 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.455 |
0.214 |
| 288 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.455 |
0.112 |
| 289 |
258480_at |
AT3G02640
|
unknown |
-0.454 |
0.278 |
| 290 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.452 |
-0.000 |
| 291 |
246505_at |
AT5G16250
|
unknown |
-0.450 |
0.371 |
| 292 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.448 |
0.153 |
| 293 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.448 |
-0.017 |
| 294 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.444 |
0.139 |
| 295 |
257794_at |
AT3G27050
|
unknown |
-0.443 |
-0.214 |
| 296 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.442 |
0.256 |
| 297 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.441 |
0.049 |
| 298 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.436 |
-0.258 |
| 299 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-0.432 |
0.168 |
| 300 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.432 |
0.037 |
| 301 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.432 |
-0.065 |
| 302 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.430 |
-0.071 |
| 303 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.428 |
0.075 |
| 304 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.428 |
-0.076 |
| 305 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.427 |
-0.047 |
| 306 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.426 |
-0.074 |
| 307 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.426 |
-0.193 |
| 308 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.425 |
-0.212 |
| 309 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-0.424 |
-0.231 |
| 310 |
267636_at |
AT2G42110
|
unknown |
-0.419 |
0.031 |
| 311 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.418 |
0.195 |
| 312 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.415 |
0.528 |
| 313 |
248205_at |
AT5G54300
|
unknown |
-0.410 |
0.016 |
| 314 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.409 |
-0.060 |
| 315 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.409 |
0.268 |
| 316 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.407 |
-0.135 |
| 317 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.403 |
-0.187 |
| 318 |
254145_at |
AT4G24700
|
unknown |
-0.402 |
-0.579 |
| 319 |
259275_at |
AT3G01060
|
unknown |
-0.399 |
-0.260 |
| 320 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.397 |
-0.454 |
| 321 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.395 |
0.574 |
| 322 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.393 |
-0.170 |
| 323 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.387 |
-0.183 |
| 324 |
250937_at |
AT5G03230
|
unknown |
-0.386 |
-0.221 |
| 325 |
246783_at |
AT5G27360
|
SFP2 |
-0.386 |
-0.199 |
| 326 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.385 |
-0.398 |
| 327 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.382 |
-0.341 |
| 328 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.381 |
-0.279 |
| 329 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.381 |
0.188 |
| 330 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.381 |
-0.149 |
| 331 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.380 |
-0.191 |
| 332 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.380 |
-0.500 |
| 333 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
-0.379 |
0.146 |
| 334 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.379 |
0.205 |
| 335 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.378 |
-0.217 |
| 336 |
256799_at |
AT3G18560
|
unknown |
-0.373 |
0.236 |
| 337 |
252089_at |
AT3G52110
|
unknown |
-0.370 |
0.090 |
| 338 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.368 |
0.204 |
| 339 |
246278_at |
AT4G37190
|
[LOCATED IN: cytosol, plasma membrane] |
-0.367 |
-0.036 |
| 340 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.366 |
-0.167 |
| 341 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.366 |
-0.312 |
| 342 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.363 |
-0.308 |
| 343 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.363 |
-0.061 |
| 344 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.362 |
0.296 |
| 345 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.361 |
-0.162 |
| 346 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.360 |
0.056 |
| 347 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.358 |
0.172 |
| 348 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.358 |
0.179 |
| 349 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.357 |
-0.537 |
| 350 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.355 |
-0.428 |
| 351 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.354 |
0.159 |
| 352 |
253978_at |
AT4G26660
|
unknown |
-0.354 |
0.197 |
| 353 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.353 |
-0.134 |
| 354 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.353 |
0.019 |
| 355 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.350 |
0.011 |
| 356 |
253814_at |
AT4G28290
|
unknown |
-0.350 |
-0.039 |
| 357 |
251010_at |
AT5G02550
|
unknown |
-0.349 |
0.222 |
| 358 |
255763_at |
AT1G16730
|
unknown |
-0.348 |
-0.273 |
| 359 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.347 |
-0.092 |
| 360 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.344 |
0.030 |
| 361 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.344 |
0.216 |
| 362 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
-0.343 |
0.048 |
| 363 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.342 |
0.222 |
| 364 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.342 |
0.061 |
| 365 |
247476_at |
AT5G62330
|
unknown |
-0.342 |
-0.009 |
| 366 |
251727_at |
AT3G56290
|
unknown |
-0.342 |
-0.286 |
| 367 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
-0.340 |
0.201 |
| 368 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.338 |
-0.530 |
| 369 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.338 |
0.001 |
| 370 |
252412_at |
AT3G47295
|
unknown |
-0.337 |
-0.064 |
| 371 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
-0.337 |
-0.063 |
| 372 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
-0.334 |
-0.143 |
| 373 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.333 |
-0.549 |
| 374 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
-0.332 |
0.088 |
| 375 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.330 |
-0.318 |
| 376 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
-0.328 |
0.004 |
| 377 |
249874_at |
AT5G23070
|
AtTK1b, TK1b, thymidine kinase 1b |
-0.328 |
0.269 |
| 378 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.327 |
0.378 |
| 379 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
-0.327 |
-0.206 |
| 380 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.326 |
-0.455 |
| 381 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.325 |
0.057 |
| 382 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.325 |
-0.270 |
| 383 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.325 |
0.238 |
| 384 |
250320_at |
AT5G12840
|
ATHAP2A, AtNFYA1, EMB2220, EMBRYO DEFECTIVE 2220, HAP2A, NF-YA1, nuclear factor Y, subunit A1, NFYA1, nuclear factor Y subunit A1 |
-0.322 |
-0.133 |
| 385 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.320 |
-0.095 |
| 386 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.320 |
0.122 |
| 387 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.319 |
-0.002 |
| 388 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.318 |
0.261 |
| 389 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.318 |
0.204 |
| 390 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.315 |
0.084 |
| 391 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.314 |
-0.297 |
| 392 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.312 |
0.356 |
| 393 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-0.310 |
-0.217 |
| 394 |
259082_at |
AT3G04820
|
[Pseudouridine synthase family protein] |
-0.307 |
-0.049 |
| 395 |
264343_at |
AT1G11850
|
unknown |
-0.307 |
0.019 |
| 396 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.307 |
-0.105 |
| 397 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.307 |
0.124 |
| 398 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.306 |
-0.060 |
| 399 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.305 |
-0.196 |
| 400 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.304 |
0.196 |
| 401 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.304 |
0.025 |