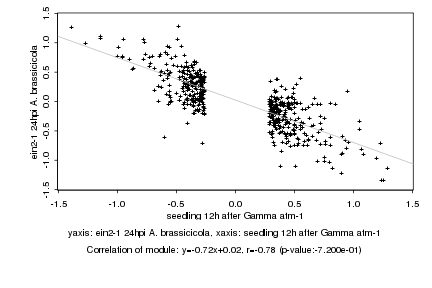
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling 12h after Gamma atm-1 |
Log2 signal ratio
ein2-1 24hpi A. brassicicola |
| 1 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.282 |
-1.131 |
| 2 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
1.255 |
-1.332 |
| 3 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
1.237 |
-1.332 |
| 4 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
1.224 |
-0.707 |
| 5 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
1.190 |
-0.965 |
| 6 |
253322_at |
AT4G33980
|
unknown |
1.084 |
-0.891 |
| 7 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
1.062 |
-0.811 |
| 8 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
1.050 |
-0.465 |
| 9 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
1.049 |
-0.326 |
| 10 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.950 |
-0.689 |
| 11 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.943 |
0.171 |
| 12 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.941 |
-0.788 |
| 13 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.932 |
-0.660 |
| 14 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.906 |
-0.584 |
| 15 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.902 |
-0.874 |
| 16 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.899 |
-1.213 |
| 17 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.896 |
-0.900 |
| 18 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.846 |
-0.045 |
| 19 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.816 |
-1.136 |
| 20 |
260688_at |
AT1G17665
|
unknown |
0.803 |
-0.741 |
| 21 |
256631_at |
AT3G28320
|
unknown |
0.801 |
-0.018 |
| 22 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.799 |
-0.601 |
| 23 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.789 |
-1.031 |
| 24 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.759 |
-0.666 |
| 25 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.758 |
-0.726 |
| 26 |
267036_at |
AT2G38465
|
unknown |
0.751 |
-1.013 |
| 27 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.747 |
-0.275 |
| 28 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.747 |
-0.940 |
| 29 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.727 |
-0.472 |
| 30 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.718 |
-0.374 |
| 31 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.718 |
-0.728 |
| 32 |
256602_at |
AT3G28310
|
unknown |
0.715 |
-0.244 |
| 33 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.714 |
-0.037 |
| 34 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.704 |
-0.514 |
| 35 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.692 |
-1.007 |
| 36 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.688 |
-0.050 |
| 37 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.681 |
-0.657 |
| 38 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.670 |
0.058 |
| 39 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.660 |
-0.402 |
| 40 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.649 |
-0.595 |
| 41 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.647 |
-0.047 |
| 42 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.644 |
-0.421 |
| 43 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.622 |
-0.279 |
| 44 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.618 |
-0.099 |
| 45 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.615 |
-0.631 |
| 46 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.615 |
-0.655 |
| 47 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.607 |
-0.738 |
| 48 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.594 |
-0.572 |
| 49 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.586 |
-0.133 |
| 50 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.582 |
-0.667 |
| 51 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
0.578 |
-0.535 |
| 52 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.574 |
-0.370 |
| 53 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.570 |
-0.740 |
| 54 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.567 |
-0.429 |
| 55 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.563 |
-0.129 |
| 56 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.555 |
-0.342 |
| 57 |
263829_at |
AT2G40435
|
unknown |
0.551 |
0.408 |
| 58 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.550 |
-0.033 |
| 59 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.548 |
-0.392 |
| 60 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.548 |
-0.470 |
| 61 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
0.533 |
-0.705 |
| 62 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.532 |
-0.643 |
| 63 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
0.527 |
-0.279 |
| 64 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.523 |
-0.619 |
| 65 |
254287_at |
AT4G22960
|
unknown |
0.520 |
-0.024 |
| 66 |
262452_at |
AT1G11210
|
unknown |
0.520 |
-0.737 |
| 67 |
252330_at |
AT3G48770
|
[DNA binding] |
0.517 |
-0.031 |
| 68 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.516 |
-0.436 |
| 69 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.512 |
0.295 |
| 70 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.505 |
-1.090 |
| 71 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.502 |
-0.310 |
| 72 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.500 |
-0.559 |
| 73 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.500 |
-0.096 |
| 74 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.498 |
0.037 |
| 75 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.496 |
-0.492 |
| 76 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
0.496 |
-0.191 |
| 77 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
0.495 |
-0.534 |
| 78 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.494 |
-0.400 |
| 79 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.494 |
-0.008 |
| 80 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.494 |
-0.739 |
| 81 |
245600_at |
AT4G14230
|
unknown |
0.494 |
-0.390 |
| 82 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.489 |
-0.460 |
| 83 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.488 |
-0.489 |
| 84 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.484 |
0.227 |
| 85 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.484 |
0.137 |
| 86 |
264314_at |
AT1G70420
|
unknown |
0.479 |
-0.040 |
| 87 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.478 |
0.071 |
| 88 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.471 |
-0.022 |
| 89 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.471 |
-0.125 |
| 90 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.470 |
0.071 |
| 91 |
261318_at |
AT1G53035
|
unknown |
0.466 |
-0.576 |
| 92 |
246929_at |
AT5G25210
|
unknown |
0.466 |
-0.756 |
| 93 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.466 |
-0.556 |
| 94 |
254659_at |
AT4G18240
|
ATSS4, ARABIDOPSIS THALIANA STARCH SYNTHASE 4, SS4, starch synthase 4, SSIV, STARCH SYNTHASE 4 |
0.466 |
-0.532 |
| 95 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.464 |
-0.297 |
| 96 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.453 |
-0.361 |
| 97 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.452 |
-0.055 |
| 98 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.450 |
-0.113 |
| 99 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.450 |
-0.085 |
| 100 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
0.448 |
-0.048 |
| 101 |
261203_at |
AT1G12845
|
unknown |
0.445 |
-0.671 |
| 102 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.445 |
-0.033 |
| 103 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.443 |
-0.102 |
| 104 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
0.443 |
-0.427 |
| 105 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.442 |
0.071 |
| 106 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
0.442 |
-0.009 |
| 107 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.439 |
-0.536 |
| 108 |
247248_at |
AT5G64560
|
ATMGT9, MGT9, magnesium transporter 9, MRS2-2 |
0.439 |
-0.084 |
| 109 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
0.438 |
-0.342 |
| 110 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.437 |
0.063 |
| 111 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.433 |
-0.334 |
| 112 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.431 |
-0.623 |
| 113 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.428 |
-0.708 |
| 114 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
0.425 |
-0.341 |
| 115 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.425 |
-0.585 |
| 116 |
265790_at |
AT2G01170
|
AtGABP, BAT1, bidirectional amino acid transporter 1, GABP, GABA permease |
0.421 |
-0.387 |
| 117 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.419 |
-0.294 |
| 118 |
262236_at |
AT1G48330
|
unknown |
0.419 |
-0.137 |
| 119 |
263515_at |
AT2G21640
|
unknown |
0.418 |
-0.268 |
| 120 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.417 |
-0.180 |
| 121 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.417 |
-0.549 |
| 122 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.416 |
0.214 |
| 123 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.415 |
-0.032 |
| 124 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.414 |
-0.531 |
| 125 |
266707_at |
AT2G03310
|
unknown |
0.413 |
-0.212 |
| 126 |
250382_at |
AT5G11580
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.411 |
-0.254 |
| 127 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
0.410 |
-0.059 |
| 128 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
0.409 |
-0.210 |
| 129 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.406 |
-0.441 |
| 130 |
247239_at |
AT5G64640
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.406 |
-0.264 |
| 131 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
0.404 |
-0.074 |
| 132 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
0.403 |
-0.246 |
| 133 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.401 |
-0.057 |
| 134 |
257421_at |
AT1G12030
|
unknown |
0.399 |
-0.294 |
| 135 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.397 |
-0.058 |
| 136 |
261921_at |
AT1G65900
|
unknown |
0.396 |
-0.013 |
| 137 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.394 |
-0.691 |
| 138 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.393 |
-0.320 |
| 139 |
245188_at |
AT1G67660
|
[Restriction endonuclease, type II-like superfamily protein] |
0.392 |
-0.506 |
| 140 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
0.387 |
-0.033 |
| 141 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.386 |
-0.849 |
| 142 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.386 |
-0.032 |
| 143 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
0.386 |
0.082 |
| 144 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.382 |
0.266 |
| 145 |
251753_at |
AT3G55760
|
unknown |
0.380 |
-0.521 |
| 146 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.380 |
-0.066 |
| 147 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.379 |
-1.098 |
| 148 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
0.379 |
-0.136 |
| 149 |
259118_at |
AT3G01310
|
[Phosphoglycerate mutase-like family protein] |
0.379 |
-0.424 |
| 150 |
253351_at |
AT4G33700
|
unknown |
0.377 |
-0.159 |
| 151 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
0.376 |
-0.040 |
| 152 |
246374_at |
AT1G51840
|
[protein kinase-related] |
0.374 |
-0.000 |
| 153 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.373 |
-0.216 |
| 154 |
246584_at |
AT5G14730
|
unknown |
0.373 |
-0.155 |
| 155 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.372 |
-0.448 |
| 156 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.367 |
-0.531 |
| 157 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.366 |
-0.343 |
| 158 |
262609_at |
AT1G13930
|
unknown |
0.366 |
-0.318 |
| 159 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
0.365 |
-0.181 |
| 160 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
0.362 |
-0.280 |
| 161 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.362 |
0.042 |
| 162 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.362 |
-0.123 |
| 163 |
267268_at |
AT2G02570
|
[nucleic acid binding] |
0.358 |
-0.306 |
| 164 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.357 |
-0.392 |
| 165 |
254834_at |
AT4G12300
|
CYP706A4, cytochrome P450, family 706, subfamily A, polypeptide 4 |
0.356 |
0.054 |
| 166 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.356 |
-0.156 |
| 167 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
0.356 |
-0.116 |
| 168 |
246829_at |
AT5G26570
|
ATGWD3, OK1, PWD, PHOSPHOGLUCAN WATER DIKINASE |
0.353 |
-0.341 |
| 169 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
0.351 |
-0.063 |
| 170 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.351 |
0.387 |
| 171 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.349 |
-0.336 |
| 172 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.349 |
-0.162 |
| 173 |
265481_at |
AT2G15960
|
unknown |
0.348 |
-0.467 |
| 174 |
253292_at |
AT4G33985
|
unknown |
0.347 |
-0.306 |
| 175 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.347 |
-0.343 |
| 176 |
259409_at |
AT1G13330
|
AHP2, Arabidopsis Hop2 homolog |
0.346 |
-0.059 |
| 177 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
0.346 |
0.385 |
| 178 |
245352_at |
AT4G15490
|
UGT84A3 |
0.343 |
0.041 |
| 179 |
252166_at |
AT3G50500
|
SNRK2-2, SNF1-RELATED PROTEIN KINASE 2-2, SNRK2.2, SNF1-related protein kinase 2.2, SPK-2-2, SNF1-RELATED PROTEIN KINASE 2-2, SRK2D |
0.343 |
-0.321 |
| 180 |
259721_at |
AT1G60890
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
0.343 |
-0.036 |
| 181 |
253733_at |
AT4G29170
|
ATMND1 |
0.343 |
-0.118 |
| 182 |
256091_at |
AT1G20693
|
HMG BETA 1, HIGH MOBILITY GROUP BETA 1, HMGB2, high mobility group B2, NFD02, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 02, NFD2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 2 |
0.342 |
-0.166 |
| 183 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.342 |
0.173 |
| 184 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.341 |
0.058 |
| 185 |
257596_at |
AT3G24760
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.340 |
-0.128 |
| 186 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
0.338 |
-0.065 |
| 187 |
250372_at |
AT5G11460
|
unknown |
0.336 |
0.009 |
| 188 |
257154_at |
AT3G27210
|
unknown |
0.336 |
-0.311 |
| 189 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.335 |
-0.433 |
| 190 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.335 |
0.156 |
| 191 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
0.335 |
-0.208 |
| 192 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.334 |
-0.083 |
| 193 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
0.334 |
-0.195 |
| 194 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
0.333 |
-0.367 |
| 195 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
0.332 |
-0.110 |
| 196 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.332 |
0.049 |
| 197 |
262533_at |
AT1G17090
|
unknown |
0.329 |
0.191 |
| 198 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.328 |
-0.128 |
| 199 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.327 |
0.083 |
| 200 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.326 |
-0.734 |
| 201 |
262722_at |
AT1G43620
|
TT15, TRANSPARENT TESTA 15, UGT80B1 |
0.326 |
-0.394 |
| 202 |
262248_at |
AT1G48370
|
YSL8, YELLOW STRIPE like 8 |
0.326 |
-0.162 |
| 203 |
247347_at |
AT5G63780
|
SHA1, shoot apical meristem arrest 1 |
0.325 |
-0.347 |
| 204 |
253553_at |
AT4G31050
|
LIP2, octanoyl- transferase |
0.325 |
-0.398 |
| 205 |
249069_at |
AT5G44010
|
unknown |
0.323 |
-0.363 |
| 206 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.321 |
-0.109 |
| 207 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
0.320 |
-0.091 |
| 208 |
249941_at |
AT5G22270
|
unknown |
0.320 |
-0.247 |
| 209 |
257611_at |
AT3G26580
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.319 |
-0.180 |
| 210 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
0.319 |
-0.319 |
| 211 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
0.318 |
-0.166 |
| 212 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.317 |
-0.682 |
| 213 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
0.317 |
-0.195 |
| 214 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.317 |
-0.200 |
| 215 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.315 |
-0.402 |
| 216 |
246525_at |
AT5G15840
|
BBX1, B-box domain protein 1, CO, CONSTANS, FG |
0.315 |
-0.311 |
| 217 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.313 |
-0.603 |
| 218 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.313 |
-0.207 |
| 219 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.311 |
-0.542 |
| 220 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.311 |
-0.012 |
| 221 |
261221_at |
AT1G19960
|
unknown |
0.311 |
-0.134 |
| 222 |
266396_at |
AT2G38790
|
unknown |
0.311 |
-0.135 |
| 223 |
254645_at |
AT4G18520
|
SEL1, SEEDLING LETHAL 1 |
0.311 |
-0.502 |
| 224 |
246155_at |
AT5G20030
|
[Plant Tudor-like RNA-binding protein] |
0.309 |
-0.148 |
| 225 |
263249_at |
AT2G31360
|
ADS2, 16:0delta9 desaturase 2, AtADS2 |
0.308 |
-0.283 |
| 226 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.306 |
-0.035 |
| 227 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.305 |
0.004 |
| 228 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.304 |
-0.034 |
| 229 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.303 |
-0.556 |
| 230 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
0.303 |
-0.087 |
| 231 |
248581_at |
AT5G49900
|
[Beta-glucosidase, GBA2 type family protein] |
0.300 |
-0.248 |
| 232 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.299 |
0.030 |
| 233 |
264588_at |
AT2G17730
|
NIP2, NEP-interacting protein 2 |
0.299 |
0.088 |
| 234 |
264096_at |
AT1G78995
|
unknown |
0.298 |
-0.157 |
| 235 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.298 |
-0.147 |
| 236 |
263065_at |
AT2G18170
|
ATMPK7, MAP kinase 7, MPK7, MAP kinase 7 |
0.296 |
-0.271 |
| 237 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
0.295 |
-0.133 |
| 238 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
0.294 |
-0.076 |
| 239 |
267378_at |
AT2G26200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.293 |
-0.352 |
| 240 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.293 |
-0.032 |
| 241 |
262503_at |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
0.293 |
-0.169 |
| 242 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.293 |
0.357 |
| 243 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.293 |
-0.357 |
| 244 |
249535_at |
AT5G38820
|
[Transmembrane amino acid transporter family protein] |
0.291 |
-0.010 |
| 245 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.289 |
-0.162 |
| 246 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.288 |
-0.391 |
| 247 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.288 |
0.012 |
| 248 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.288 |
-0.356 |
| 249 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.287 |
-0.040 |
| 250 |
254764_at |
AT4G13250
|
NYC1, NON-YELLOW COLORING 1 |
0.286 |
0.036 |
| 251 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
0.286 |
-0.522 |
| 252 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.285 |
-0.249 |
| 253 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
0.285 |
-0.193 |
| 254 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-1.396 |
1.273 |
| 255 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-1.279 |
1.001 |
| 256 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-1.146 |
1.117 |
| 257 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-1.145 |
1.084 |
| 258 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-1.006 |
0.778 |
| 259 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.996 |
0.934 |
| 260 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.961 |
0.763 |
| 261 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.959 |
0.779 |
| 262 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.950 |
1.056 |
| 263 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.901 |
0.729 |
| 264 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.879 |
0.560 |
| 265 |
264636_at |
AT1G65490
|
unknown |
-0.866 |
0.579 |
| 266 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.785 |
0.718 |
| 267 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.785 |
1.068 |
| 268 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.773 |
1.015 |
| 269 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.769 |
0.804 |
| 270 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.737 |
0.761 |
| 271 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.729 |
0.610 |
| 272 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.725 |
0.650 |
| 273 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.710 |
0.781 |
| 274 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.691 |
0.204 |
| 275 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
-0.689 |
0.576 |
| 276 |
261658_at |
AT1G50040
|
unknown |
-0.659 |
0.014 |
| 277 |
259373_at |
AT1G69160
|
unknown |
-0.655 |
0.265 |
| 278 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.653 |
0.269 |
| 279 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.644 |
0.769 |
| 280 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.642 |
0.459 |
| 281 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.642 |
0.498 |
| 282 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.638 |
0.806 |
| 283 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.631 |
0.525 |
| 284 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.628 |
0.254 |
| 285 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.618 |
0.368 |
| 286 |
247474_at |
AT5G62280
|
unknown |
-0.607 |
-0.602 |
| 287 |
249932_at |
AT5G22390
|
unknown |
-0.602 |
0.489 |
| 288 |
256096_at |
AT1G13650
|
unknown |
-0.596 |
0.850 |
| 289 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.590 |
0.134 |
| 290 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
-0.588 |
0.647 |
| 291 |
248205_at |
AT5G54300
|
unknown |
-0.581 |
0.446 |
| 292 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.578 |
0.802 |
| 293 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.577 |
0.403 |
| 294 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.576 |
0.539 |
| 295 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.576 |
0.946 |
| 296 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.575 |
0.453 |
| 297 |
261567_at |
AT1G33055
|
unknown |
-0.570 |
-0.050 |
| 298 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.565 |
0.114 |
| 299 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.564 |
0.284 |
| 300 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.563 |
0.480 |
| 301 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.561 |
0.180 |
| 302 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.560 |
0.926 |
| 303 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.556 |
0.002 |
| 304 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.554 |
0.507 |
| 305 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.554 |
-0.015 |
| 306 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.553 |
0.069 |
| 307 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.550 |
0.735 |
| 308 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.547 |
0.017 |
| 309 |
251727_at |
AT3G56290
|
unknown |
-0.533 |
0.621 |
| 310 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.512 |
0.774 |
| 311 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.503 |
0.153 |
| 312 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.495 |
1.059 |
| 313 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.494 |
0.610 |
| 314 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.488 |
1.282 |
| 315 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
-0.482 |
0.521 |
| 316 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.482 |
0.409 |
| 317 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.480 |
0.024 |
| 318 |
253919_at |
AT4G27350
|
unknown |
-0.478 |
0.024 |
| 319 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.478 |
0.377 |
| 320 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.470 |
0.087 |
| 321 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.465 |
0.946 |
| 322 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.459 |
0.611 |
| 323 |
262879_at |
AT1G64860
|
RPOD1, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, sigma factor A, SIGB |
-0.454 |
0.505 |
| 324 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.450 |
0.234 |
| 325 |
252882_at |
AT4G39675
|
unknown |
-0.450 |
0.427 |
| 326 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.447 |
0.266 |
| 327 |
266222_at |
AT2G28780
|
unknown |
-0.445 |
-0.035 |
| 328 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.445 |
0.173 |
| 329 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.443 |
0.543 |
| 330 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
-0.442 |
0.354 |
| 331 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.440 |
0.392 |
| 332 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-0.437 |
0.312 |
| 333 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
-0.435 |
0.793 |
| 334 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.435 |
0.489 |
| 335 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.434 |
0.592 |
| 336 |
245353_at |
AT4G16000
|
unknown |
-0.433 |
-0.110 |
| 337 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.432 |
-0.006 |
| 338 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.430 |
0.293 |
| 339 |
262786_at |
AT1G10740
|
[alpha/beta-Hydrolases superfamily protein] |
-0.428 |
0.260 |
| 340 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.425 |
0.126 |
| 341 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.423 |
0.671 |
| 342 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.420 |
0.041 |
| 343 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.418 |
0.613 |
| 344 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.418 |
0.317 |
| 345 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.417 |
0.383 |
| 346 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.416 |
0.600 |
| 347 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.414 |
0.474 |
| 348 |
246552_at |
AT5G15420
|
unknown |
-0.413 |
0.027 |
| 349 |
250327_at |
AT5G12050
|
unknown |
-0.413 |
-0.363 |
| 350 |
261171_at |
AT1G04880
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.412 |
-0.038 |
| 351 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.412 |
0.199 |
| 352 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.412 |
0.236 |
| 353 |
252412_at |
AT3G47295
|
unknown |
-0.412 |
0.314 |
| 354 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.410 |
0.542 |
| 355 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.408 |
0.250 |
| 356 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.407 |
0.545 |
| 357 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.407 |
0.054 |
| 358 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
-0.406 |
0.286 |
| 359 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.405 |
0.592 |
| 360 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.400 |
0.319 |
| 361 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
-0.396 |
0.423 |
| 362 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.393 |
0.522 |
| 363 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.391 |
0.211 |
| 364 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.391 |
0.113 |
| 365 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
-0.388 |
0.110 |
| 366 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.388 |
0.352 |
| 367 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.387 |
-0.035 |
| 368 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.387 |
0.117 |
| 369 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.384 |
0.475 |
| 370 |
250972_at |
AT5G02840
|
LCL1, LHY/CCA1-like 1 |
-0.383 |
0.322 |
| 371 |
246783_at |
AT5G27360
|
SFP2 |
-0.380 |
0.296 |
| 372 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.379 |
0.311 |
| 373 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.376 |
0.350 |
| 374 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.376 |
0.625 |
| 375 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.375 |
0.437 |
| 376 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.375 |
0.037 |
| 377 |
254145_at |
AT4G24700
|
unknown |
-0.374 |
0.619 |
| 378 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-0.373 |
0.451 |
| 379 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.372 |
0.185 |
| 380 |
261667_at |
AT1G18460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.371 |
0.303 |
| 381 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.370 |
0.334 |
| 382 |
263632_at |
AT2G04795
|
unknown |
-0.369 |
0.271 |
| 383 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
-0.367 |
0.407 |
| 384 |
267063_at |
AT2G41120
|
unknown |
-0.367 |
0.340 |
| 385 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.367 |
0.033 |
| 386 |
249122_at |
AT5G43850
|
ARD4, ATARD4 |
-0.367 |
0.314 |
| 387 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.366 |
0.046 |
| 388 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.366 |
0.018 |
| 389 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.365 |
0.587 |
| 390 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.364 |
0.013 |
| 391 |
260137_at |
AT1G66330
|
[senescence-associated family protein] |
-0.364 |
0.398 |
| 392 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.362 |
-0.195 |
| 393 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.361 |
0.396 |
| 394 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.360 |
-0.054 |
| 395 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.360 |
0.007 |
| 396 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.359 |
0.086 |
| 397 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.358 |
-0.004 |
| 398 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.357 |
0.066 |
| 399 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.357 |
0.532 |
| 400 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.357 |
-0.073 |
| 401 |
253394_at |
AT4G32770
|
ATSDX1, SUCROSE EXPORT DEFECTIVE 1, VTE1, VITAMIN E DEFICIENT 1 |
-0.355 |
0.410 |
| 402 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.355 |
0.691 |
| 403 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.354 |
-0.193 |
| 404 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.352 |
0.638 |
| 405 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.350 |
0.671 |
| 406 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
-0.350 |
0.127 |
| 407 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.348 |
0.275 |
| 408 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.347 |
-0.138 |
| 409 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.347 |
0.292 |
| 410 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.346 |
-0.008 |
| 411 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.346 |
0.031 |
| 412 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.346 |
0.487 |
| 413 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.345 |
0.258 |
| 414 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.343 |
0.313 |
| 415 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.342 |
0.238 |
| 416 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.339 |
0.056 |
| 417 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.338 |
0.336 |
| 418 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.338 |
0.207 |
| 419 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.337 |
0.431 |
| 420 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.337 |
0.673 |
| 421 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.333 |
0.437 |
| 422 |
256674_at |
AT3G52360
|
unknown |
-0.330 |
0.152 |
| 423 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.329 |
0.340 |
| 424 |
247476_at |
AT5G62330
|
unknown |
-0.328 |
0.039 |
| 425 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.325 |
0.502 |
| 426 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
-0.325 |
0.436 |
| 427 |
264521_at |
AT1G10020
|
unknown |
-0.324 |
0.050 |
| 428 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.323 |
-0.129 |
| 429 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.323 |
0.145 |
| 430 |
256861_at |
AT3G23920
|
AtBAM1, BAM1, beta-amylase 1, BMY7, BETA-AMYLASE 7, TR-BAMY |
-0.321 |
0.468 |
| 431 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.320 |
0.028 |
| 432 |
264040_at |
AT2G03730
|
ACR5, ACT domain repeat 5 |
-0.319 |
0.342 |
| 433 |
258729_at |
AT3G11900
|
ANT1, aromatic and neutral transporter 1 |
-0.317 |
0.266 |
| 434 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.317 |
-0.167 |
| 435 |
257076_at |
AT3G19680
|
unknown |
-0.316 |
-0.062 |
| 436 |
260875_at |
AT1G21410
|
SKP2A |
-0.311 |
0.533 |
| 437 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.311 |
0.349 |
| 438 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.310 |
0.011 |
| 439 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.309 |
0.148 |
| 440 |
260696_at |
AT1G32520
|
unknown |
-0.309 |
0.421 |
| 441 |
262388_at |
AT1G49320
|
unknown |
-0.307 |
-0.127 |
| 442 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
-0.307 |
0.097 |
| 443 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.306 |
0.053 |
| 444 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.305 |
-0.045 |
| 445 |
263805_at |
AT2G40400
|
unknown |
-0.303 |
0.395 |
| 446 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.303 |
0.416 |
| 447 |
253039_at |
AT4G37760
|
SQE3, squalene epoxidase 3 |
-0.302 |
0.425 |
| 448 |
259275_at |
AT3G01060
|
unknown |
-0.302 |
0.422 |
| 449 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.302 |
0.040 |
| 450 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.301 |
0.472 |
| 451 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.299 |
0.260 |
| 452 |
245984_at |
AT5G13090
|
unknown |
-0.298 |
0.140 |
| 453 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.297 |
0.415 |
| 454 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.297 |
0.428 |
| 455 |
258196_at |
AT3G13980
|
unknown |
-0.297 |
0.095 |
| 456 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.297 |
0.542 |
| 457 |
266717_at |
AT2G46735
|
unknown |
-0.297 |
0.214 |
| 458 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
-0.295 |
0.302 |
| 459 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.294 |
0.070 |
| 460 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.294 |
0.431 |
| 461 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.294 |
0.103 |
| 462 |
251797_at |
AT3G55560
|
AGF2, AT-hook protein of GA feedback 2, AHL15, AT-hook motif nuclear-localized protein 15 |
-0.293 |
0.006 |
| 463 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.293 |
0.358 |
| 464 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
-0.292 |
0.071 |
| 465 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.292 |
0.160 |
| 466 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.290 |
0.389 |
| 467 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
-0.290 |
0.270 |
| 468 |
250008_at |
AT5G18630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.290 |
0.145 |
| 469 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.290 |
0.301 |
| 470 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.290 |
0.169 |
| 471 |
245981_at |
AT5G13100
|
unknown |
-0.289 |
0.051 |
| 472 |
253814_at |
AT4G28290
|
unknown |
-0.289 |
0.369 |
| 473 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.288 |
0.330 |
| 474 |
266864_at |
AT2G26800
|
[Aldolase superfamily protein] |
-0.288 |
0.280 |
| 475 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
-0.288 |
0.317 |
| 476 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.287 |
0.239 |
| 477 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.286 |
-0.000 |
| 478 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
-0.285 |
0.333 |
| 479 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.284 |
-0.699 |
| 480 |
259484_at |
AT1G12580
|
PEPKR1, phosphoenolpyruvate carboxylase-related kinase 1 |
-0.284 |
0.181 |
| 481 |
264512_at |
AT1G09575
|
unknown |
-0.283 |
-0.189 |
| 482 |
246926_at |
AT5G25240
|
unknown |
-0.282 |
0.041 |
| 483 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.282 |
0.079 |
| 484 |
246999_at |
AT5G67440
|
MEL2, MAB4/ENP/NPY1-LIKE 2, NPY3, NAKED PINS IN YUC MUTANTS 3 |
-0.280 |
0.185 |
| 485 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.279 |
-0.038 |
| 486 |
260543_at |
AT2G43330
|
ATINT1, INOSITOL TRANSPORTER 1, INT1, inositol transporter 1 |
-0.278 |
0.131 |
| 487 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.277 |
0.154 |
| 488 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.275 |
0.376 |
| 489 |
250223_at |
AT5G14070
|
ROXY2 |
-0.273 |
-0.141 |
| 490 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
-0.272 |
-0.099 |
| 491 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.272 |
0.054 |
| 492 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.272 |
0.415 |
| 493 |
257855_at |
AT3G13040
|
[myb-like HTH transcriptional regulator family protein] |
-0.272 |
0.358 |
| 494 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.271 |
-0.069 |
| 495 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.271 |
0.397 |
| 496 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
-0.270 |
0.180 |
| 497 |
248080_at |
AT5G55380
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.269 |
0.327 |
| 498 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.269 |
-0.113 |
| 499 |
257710_at |
AT3G27350
|
unknown |
-0.269 |
0.443 |
| 500 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.268 |
-0.213 |
| 501 |
256456_at |
AT1G75180
|
[Erythronate-4-phosphate dehydrogenase family protein] |
-0.268 |
0.421 |
| 502 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
-0.268 |
0.219 |
| 503 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.267 |
-0.188 |
| 504 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
-0.267 |
0.511 |
| 505 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.267 |
0.202 |