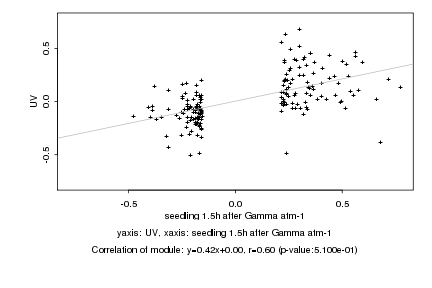
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling 1.5h after Gamma atm-1 |
Log2 signal ratio
UV |
| 1 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.771 |
0.141 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.713 |
0.208 |
| 3 |
246001_at |
AT5G20790
|
unknown |
0.677 |
-0.384 |
| 4 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.658 |
0.023 |
| 5 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.594 |
0.371 |
| 6 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.575 |
0.109 |
| 7 |
253044_at |
AT4G37290
|
unknown |
0.562 |
0.424 |
| 8 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.561 |
0.461 |
| 9 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.550 |
0.057 |
| 10 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.537 |
0.099 |
| 11 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.527 |
0.240 |
| 12 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.517 |
0.351 |
| 13 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.512 |
-0.058 |
| 14 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.499 |
0.381 |
| 15 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
0.492 |
0.004 |
| 16 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.489 |
-0.005 |
| 17 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.481 |
0.173 |
| 18 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.467 |
0.059 |
| 19 |
245082_at |
AT2G23270
|
unknown |
0.460 |
0.236 |
| 20 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.438 |
0.437 |
| 21 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.438 |
0.221 |
| 22 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.422 |
0.021 |
| 23 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.403 |
0.319 |
| 24 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.402 |
0.174 |
| 25 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
0.399 |
0.056 |
| 26 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.381 |
0.028 |
| 27 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.366 |
0.374 |
| 28 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.363 |
0.269 |
| 29 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.362 |
0.118 |
| 30 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.360 |
0.147 |
| 31 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.351 |
0.060 |
| 32 |
258203_at |
AT3G13950
|
unknown |
0.350 |
0.455 |
| 33 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.349 |
0.456 |
| 34 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.343 |
0.124 |
| 35 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.339 |
0.132 |
| 36 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.337 |
-0.074 |
| 37 |
266017_at |
AT2G18690
|
unknown |
0.334 |
0.185 |
| 38 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.332 |
0.348 |
| 39 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.331 |
0.079 |
| 40 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.330 |
-0.052 |
| 41 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.324 |
0.000 |
| 42 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.320 |
0.414 |
| 43 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.317 |
0.248 |
| 44 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.316 |
-0.113 |
| 45 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.316 |
0.404 |
| 46 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.301 |
-0.058 |
| 47 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.299 |
0.254 |
| 48 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.297 |
0.326 |
| 49 |
252661_at |
AT3G44450
|
unknown |
0.296 |
0.680 |
| 50 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.296 |
0.520 |
| 51 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.287 |
-0.019 |
| 52 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.285 |
0.392 |
| 53 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.281 |
-0.065 |
| 54 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.279 |
0.079 |
| 55 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.276 |
0.062 |
| 56 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.275 |
0.401 |
| 57 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.266 |
0.212 |
| 58 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
0.266 |
-0.062 |
| 59 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.265 |
-0.017 |
| 60 |
252010_at |
AT3G52740
|
unknown |
0.256 |
0.315 |
| 61 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.255 |
0.496 |
| 62 |
259479_at |
AT1G19020
|
unknown |
0.254 |
0.174 |
| 63 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.251 |
0.297 |
| 64 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.248 |
0.138 |
| 65 |
250372_at |
AT5G11460
|
unknown |
0.244 |
0.048 |
| 66 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.241 |
0.206 |
| 67 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.239 |
0.119 |
| 68 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.239 |
0.260 |
| 69 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.238 |
0.082 |
| 70 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.237 |
-0.028 |
| 71 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.236 |
-0.481 |
| 72 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.235 |
0.072 |
| 73 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.231 |
0.634 |
| 74 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.230 |
0.215 |
| 75 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.229 |
0.193 |
| 76 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.228 |
0.204 |
| 77 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.228 |
-0.021 |
| 78 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.227 |
0.370 |
| 79 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.226 |
0.387 |
| 80 |
247769_at |
AT5G58910
|
LAC16, laccase 16 |
0.226 |
-0.005 |
| 81 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.225 |
0.155 |
| 82 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
0.225 |
0.002 |
| 83 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.225 |
0.089 |
| 84 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.224 |
0.028 |
| 85 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
0.224 |
-0.037 |
| 86 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
0.223 |
-0.030 |
| 87 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
0.215 |
-0.088 |
| 88 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.214 |
0.088 |
| 89 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.213 |
0.562 |
| 90 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
0.213 |
-0.015 |
| 91 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.211 |
0.044 |
| 92 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.481 |
-0.139 |
| 93 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.411 |
-0.056 |
| 94 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.399 |
-0.143 |
| 95 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.393 |
-0.039 |
| 96 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.390 |
-0.083 |
| 97 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.380 |
0.149 |
| 98 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.374 |
-0.164 |
| 99 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.348 |
-0.148 |
| 100 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.325 |
-0.328 |
| 101 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.318 |
-0.432 |
| 102 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.317 |
-0.075 |
| 103 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.315 |
0.111 |
| 104 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.277 |
-0.123 |
| 105 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.262 |
-0.159 |
| 106 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
-0.253 |
-0.101 |
| 107 |
254193_at |
AT4G23870
|
unknown |
-0.253 |
-0.317 |
| 108 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.249 |
0.162 |
| 109 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.248 |
0.049 |
| 110 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.248 |
0.031 |
| 111 |
252089_at |
AT3G52110
|
unknown |
-0.247 |
-0.104 |
| 112 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.242 |
-0.069 |
| 113 |
259546_at |
AT1G35350
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.236 |
0.076 |
| 114 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.233 |
-0.237 |
| 115 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.231 |
0.170 |
| 116 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
-0.229 |
-0.069 |
| 117 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.228 |
0.010 |
| 118 |
267636_at |
AT2G42110
|
unknown |
-0.228 |
-0.046 |
| 119 |
251299_at |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.227 |
-0.194 |
| 120 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.227 |
-0.022 |
| 121 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
-0.226 |
-0.141 |
| 122 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.216 |
-0.064 |
| 123 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.216 |
-0.302 |
| 124 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.215 |
-0.040 |
| 125 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.214 |
-0.056 |
| 126 |
266956_at |
AT2G34510
|
unknown |
-0.214 |
-0.176 |
| 127 |
250936_at |
AT5G03120
|
unknown |
-0.212 |
-0.502 |
| 128 |
252215_at |
AT3G50240
|
KICP-02 |
-0.207 |
-0.142 |
| 129 |
257547_at |
AT3G13000
|
unknown |
-0.206 |
-0.279 |
| 130 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.200 |
-0.073 |
| 131 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.199 |
-0.102 |
| 132 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.198 |
-0.165 |
| 133 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
-0.197 |
0.021 |
| 134 |
261292_at |
AT1G36940
|
unknown |
-0.190 |
-0.081 |
| 135 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.189 |
-0.194 |
| 136 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.189 |
-0.216 |
| 137 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.189 |
-0.153 |
| 138 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.188 |
-0.096 |
| 139 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.187 |
-0.212 |
| 140 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.187 |
-0.199 |
| 141 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.186 |
-0.044 |
| 142 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.186 |
0.085 |
| 143 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.185 |
0.155 |
| 144 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
-0.185 |
-0.068 |
| 145 |
258544_at |
AT3G07040
|
RPM1, RESISTANCE TO P. SYRINGAE PV MACULICOLA 1, RPS3, RESISTANCE TO PSEUDOMONAS SYRINGAE 3 |
-0.184 |
-0.053 |
| 146 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
-0.183 |
0.060 |
| 147 |
264636_at |
AT1G65490
|
unknown |
-0.180 |
-0.147 |
| 148 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.180 |
-0.058 |
| 149 |
261405_at |
AT1G18740
|
unknown |
-0.180 |
-0.021 |
| 150 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.179 |
-0.189 |
| 151 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.178 |
-0.314 |
| 152 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.176 |
-0.112 |
| 153 |
267209_at |
AT2G30930
|
unknown |
-0.174 |
-0.143 |
| 154 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
-0.174 |
-0.162 |
| 155 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.172 |
-0.145 |
| 156 |
261026_at |
AT1G01240
|
unknown |
-0.172 |
0.063 |
| 157 |
261791_at |
AT1G16170
|
unknown |
-0.171 |
-0.232 |
| 158 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.170 |
-0.480 |
| 159 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.169 |
-0.042 |
| 160 |
258530_at |
AT3G06840
|
unknown |
-0.169 |
-0.220 |
| 161 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.166 |
-0.200 |
| 162 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.166 |
-0.116 |
| 163 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.165 |
-0.066 |
| 164 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
-0.165 |
-0.061 |
| 165 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.164 |
-0.080 |
| 166 |
250877_at |
AT5G04040
|
SDP1, SUGAR-DEPENDENT1 |
-0.164 |
0.045 |
| 167 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.164 |
0.021 |
| 168 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.164 |
-0.105 |
| 169 |
256096_at |
AT1G13650
|
unknown |
-0.164 |
-0.003 |
| 170 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.162 |
-0.332 |
| 171 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.162 |
-0.164 |
| 172 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.162 |
0.200 |
| 173 |
257140_at |
AT3G28910
|
ATMYB30, MYB30, myb domain protein 30 |
-0.162 |
-0.253 |
| 174 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.160 |
-0.263 |
| 175 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.160 |
0.063 |
| 176 |
251413_at |
AT3G60320
|
unknown |
-0.159 |
-0.096 |
| 177 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.159 |
-0.110 |
| 178 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.159 |
-0.088 |
| 179 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
-0.159 |
0.024 |
| 180 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.159 |
-0.077 |
| 181 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
-0.158 |
-0.132 |