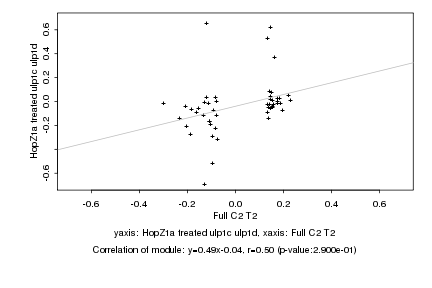
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Full C2 T2 |
Log2 signal ratio
HopZ1a treated ulp1c ulp1d |
| 1 |
256588_at |
AT3G28790
|
unknown |
0.228 |
0.016 |
| 2 |
263587_at |
AT2G25340
|
ATVAMP712, vesicle-associated membrane protein 712, VAMP712, VAMP712, vesicle-associated membrane protein 712 |
0.217 |
0.053 |
| 3 |
257305_at |
AT3G30250
|
unknown |
0.196 |
-0.069 |
| 4 |
250785_at |
AT5G05510
|
[Mad3/BUB1 homology region 1] |
0.187 |
-0.013 |
| 5 |
263045_at |
AT2G05370
|
unknown |
0.182 |
0.029 |
| 6 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
0.175 |
0.003 |
| 7 |
265064_at |
AT1G61630
|
ATENT7, ENT7, equilibrative nucleoside transporter 7 |
0.175 |
0.030 |
| 8 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
0.175 |
-0.016 |
| 9 |
251859_at |
AT3G54680
|
[proteophosphoglycan-related] |
0.159 |
0.372 |
| 10 |
260630_at |
AT1G62340
|
ALE, ABNORMAL LEAF-SHAPE, ALE1, ABNORMAL LEAF-SHAPE 1 |
0.157 |
-0.019 |
| 11 |
251019_at |
AT5G02420
|
unknown |
0.153 |
-0.044 |
| 12 |
252962_at |
AT4G38780
|
[Pre-mRNA-processing-splicing factor] |
0.151 |
0.009 |
| 13 |
255712_at |
AT4G00280
|
unknown |
0.151 |
-0.038 |
| 14 |
267438_at |
AT2G19050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.150 |
-0.044 |
| 15 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
0.149 |
0.083 |
| 16 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.143 |
0.022 |
| 17 |
246788_at |
AT5G27580
|
AGL89, AGAMOUS-like 89 |
0.143 |
0.044 |
| 18 |
250782_at |
AT5G05490
|
AtREC8, DIF1, DETERMINATE, INFERTILE 1, REC8, SYN1, SYNAPTIC 1 |
0.142 |
-0.052 |
| 19 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.142 |
0.621 |
| 20 |
251239_at |
AT3G62440
|
SAF1, secondary wall thickening-associated F-box 1 |
0.140 |
0.084 |
| 21 |
251504_at |
AT3G59030
|
ATTT12, A. THALIANA TRANSPARENT TESTA, TT12, TRANSPARENT TESTA 12 |
0.138 |
-0.022 |
| 22 |
256205_at |
AT1G50890
|
[ARM repeat superfamily protein] |
0.137 |
-0.140 |
| 23 |
250241_at |
AT5G13600
|
[Phototropic-responsive NPH3 family protein] |
0.136 |
-0.045 |
| 24 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.132 |
0.534 |
| 25 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
0.131 |
-0.018 |
| 26 |
259496_at |
AT1G15900
|
unknown |
0.130 |
-0.087 |
| 27 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.301 |
-0.009 |
| 28 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
-0.234 |
-0.138 |
| 29 |
251900_at |
AT3G54430
|
SRS6, SHI-related sequence 6 |
-0.210 |
-0.037 |
| 30 |
266187_at |
AT2G38970
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.205 |
-0.202 |
| 31 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.190 |
-0.273 |
| 32 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.186 |
-0.063 |
| 33 |
255888_at |
AT1G20300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.164 |
-0.086 |
| 34 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.155 |
-0.053 |
| 35 |
260316_at |
AT1G63810
|
unknown |
-0.135 |
-0.116 |
| 36 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
-0.132 |
-0.004 |
| 37 |
247474_at |
AT5G62280
|
unknown |
-0.129 |
-0.686 |
| 38 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.122 |
0.038 |
| 39 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
-0.121 |
0.653 |
| 40 |
246094_at |
AT5G19300
|
unknown |
-0.114 |
-0.016 |
| 41 |
249462_at |
AT5G39680
|
EMB2744, EMBRYO DEFECTIVE 2744 |
-0.109 |
-0.162 |
| 42 |
263843_at |
AT2G37020
|
[Translin family protein] |
-0.109 |
-0.162 |
| 43 |
254320_at |
AT4G22580
|
[Exostosin family protein] |
-0.105 |
-0.187 |
| 44 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.097 |
-0.285 |
| 45 |
252073_at |
AT3G51750
|
unknown |
-0.096 |
-0.515 |
| 46 |
252990_at |
AT4G38440
|
IYO, MINIYO |
-0.092 |
-0.073 |
| 47 |
264932_at |
AT1G61240
|
unknown |
-0.087 |
-0.219 |
| 48 |
257483_at |
AT1G49620
|
AtKRP7, ICK5, ICN6, KRP7, KIP-RELATED PROTEIN 7 |
-0.086 |
0.042 |
| 49 |
255270_at |
AT4G05250
|
[Ubiquitin-like superfamily protein] |
-0.081 |
0.007 |
| 50 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.079 |
-0.114 |
| 51 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
-0.077 |
-0.314 |