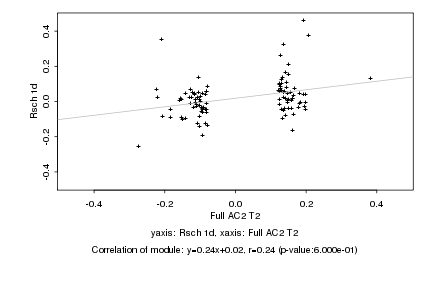
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Full AC2 T2 |
Log2 signal ratio
Rsch 1d |
| 1 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.380 |
0.132 |
| 2 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.206 |
0.378 |
| 3 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.198 |
0.043 |
| 4 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.196 |
-0.041 |
| 5 |
245158_at |
AT2G33130
|
RALF18, RALFL18, ralf-like 18 |
0.196 |
-0.004 |
| 6 |
266109_at |
AT2G37890
|
[Mitochondrial substrate carrier family protein] |
0.195 |
-0.023 |
| 7 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.192 |
0.466 |
| 8 |
245107_at |
AT2G41690
|
AT-HSFB3, heat shock transcription factor B3, HSFB3, HEAT SHOCK TRANSCRIPTION FACTOR B3, HSFB3, heat shock transcription factor B3 |
0.190 |
0.044 |
| 9 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.184 |
-0.004 |
| 10 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
0.179 |
0.051 |
| 11 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.179 |
-0.009 |
| 12 |
257201_at |
AT3G23740
|
unknown |
0.176 |
-0.032 |
| 13 |
260122_at |
AT1G33900
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.166 |
0.078 |
| 14 |
248004_at |
AT5G56230
|
PRA1.G2, prenylated RAB acceptor 1.G2 |
0.163 |
-0.073 |
| 15 |
245239_at |
AT4G25530
|
FWA, FLOWERING WAGENINGEN, HDG6, HOMEODOMAIN GLABROUS 6 |
0.162 |
0.035 |
| 16 |
249352_at |
AT5G40430
|
AtMYB22, myb domain protein 22, MYB22, myb domain protein 22 |
0.161 |
0.018 |
| 17 |
263580_at |
AT2G17140
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.160 |
0.018 |
| 18 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.160 |
-0.162 |
| 19 |
252043_at |
AT3G52390
|
[TatD related DNase] |
0.158 |
0.007 |
| 20 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.156 |
-0.039 |
| 21 |
251252_at |
AT3G62230
|
DAF1, DUO1-activated F-box 1 |
0.153 |
0.054 |
| 22 |
263476_at |
AT2G31870
|
PARG1, poly(ADP-ribose) glycohydrolase 1, TEJ, Sanskrit for 'bright' |
0.150 |
-0.038 |
| 23 |
247515_at |
AT5G61740
|
ABCA10, ATP-binding cassette A10, ATATH14, ARABIDOPSIS THALIANA ABC2 HOMOLOG 14, ATH14, ABC2 homolog 14 |
0.150 |
0.017 |
| 24 |
257331_at |
ATMG01330
|
ORF107H |
0.149 |
0.155 |
| 25 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
0.148 |
0.212 |
| 26 |
245073_at |
AT2G23210
|
[UDP-Glycosyltransferase superfamily protein] |
0.147 |
-0.003 |
| 27 |
253718_at |
AT4G29450
|
[Leucine-rich repeat protein kinase family protein] |
0.145 |
0.051 |
| 28 |
247927_at |
AT5G57310
|
unknown |
0.145 |
0.011 |
| 29 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
0.143 |
0.113 |
| 30 |
260233_at |
AT1G74550
|
CYP98A9, cytochrome P450, family 98, subfamily A, polypeptide 9 |
0.142 |
0.085 |
| 31 |
266618_at |
AT2G35480
|
unknown |
0.141 |
-0.078 |
| 32 |
262674_at |
AT1G75910
|
EXL4, extracellular lipase 4 |
0.140 |
0.021 |
| 33 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
0.139 |
0.168 |
| 34 |
245466_at |
AT4G16600
|
PGSIP8, plant glycogenin-like starch initiation protein 8 |
0.137 |
0.061 |
| 35 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.137 |
-0.037 |
| 36 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
0.136 |
0.328 |
| 37 |
248069_at |
AT5G55650
|
unknown |
0.134 |
-0.047 |
| 38 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.134 |
0.024 |
| 39 |
250342_at |
AT5G11870
|
[Alkaline phytoceramidase (aPHC)] |
0.134 |
0.052 |
| 40 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.133 |
-0.094 |
| 41 |
254917_at |
AT4G11350
|
unknown |
0.131 |
0.138 |
| 42 |
249547_at |
AT5G38160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.130 |
0.066 |
| 43 |
260859_at |
AT1G43780
|
scpl44, serine carboxypeptidase-like 44 |
0.129 |
-0.045 |
| 44 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
0.129 |
0.067 |
| 45 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
0.128 |
0.129 |
| 46 |
265176_at |
AT1G23520
|
unknown |
0.128 |
0.107 |
| 47 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
0.126 |
0.088 |
| 48 |
267604_at |
AT2G32930
|
ZFN2, zinc finger nuclease 2 |
0.125 |
0.262 |
| 49 |
253222_at |
AT4G34850
|
LAP5, LESS ADHESIVE POLLEN 5, PSKB, polyketide synthase B |
0.125 |
0.076 |
| 50 |
257026_at |
AT3G19200
|
unknown |
0.124 |
0.107 |
| 51 |
262765_at |
AT1G13150
|
CYP86C4, cytochrome P450, family 86, subfamily C, polypeptide 4 |
0.123 |
0.013 |
| 52 |
248341_at |
AT5G52220
|
unknown |
0.123 |
-0.013 |
| 53 |
253413_at |
AT4G33020
|
ATZIP9, ZIP9 |
0.123 |
0.058 |
| 54 |
260355_at |
AT1G69180
|
CRC, CRABS CLAW |
0.122 |
0.101 |
| 55 |
247059_at |
AT5G66690
|
UGT72E2 |
0.121 |
0.066 |
| 56 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
-0.276 |
-0.254 |
| 57 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.225 |
0.072 |
| 58 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
-0.223 |
0.025 |
| 59 |
262719_at |
AT1G43590
|
unknown |
-0.211 |
0.355 |
| 60 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.208 |
-0.085 |
| 61 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
-0.186 |
-0.041 |
| 62 |
251095_at |
AT5G01510
|
RUS5, ROOT UV-B SENSITIVE 5 |
-0.186 |
-0.090 |
| 63 |
267271_at |
AT2G02540
|
ATHB21, homeobox protein 21, HB21, homeobox protein 21, ZFHD4, ZINC FINGER HOMEODOMAIN 4, ZHD3, ZINC FINGER HOMEODOMAIN 3 |
-0.161 |
0.007 |
| 64 |
252734_at |
AT3G43160
|
MEE38, maternal effect embryo arrest 38 |
-0.157 |
0.022 |
| 65 |
253320_at |
AT4G34020
|
AtDJ1C, DJ-1 homolog C, DJ-1c, DJ1C, DJ-1 homolog C |
-0.153 |
-0.089 |
| 66 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.153 |
0.016 |
| 67 |
248299_at |
AT5G53080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.150 |
-0.099 |
| 68 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.142 |
0.049 |
| 69 |
245149_at |
AT2G47620
|
ATSWI3A, SWITCH/sucrose nonfermenting 3A, CHB1, SWI3A, SWITCH/sucrose nonfermenting 3A |
-0.142 |
-0.093 |
| 70 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
-0.131 |
0.023 |
| 71 |
249951_at |
AT5G18930
|
BUD2, BUSHY AND DWARF 2, SAMDC4 |
-0.129 |
-0.011 |
| 72 |
255518_at |
AT4G02300
|
PME39, pectin methylesterase 39 |
-0.128 |
0.069 |
| 73 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.125 |
0.023 |
| 74 |
258961_at |
AT3G10580
|
[Homeodomain-like superfamily protein] |
-0.122 |
0.053 |
| 75 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.119 |
-0.030 |
| 76 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.118 |
0.042 |
| 77 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.116 |
0.049 |
| 78 |
252918_at |
AT4G38980
|
unknown |
-0.114 |
0.015 |
| 79 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
-0.114 |
-0.000 |
| 80 |
263230_at |
AT1G05670
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.113 |
-0.023 |
| 81 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.113 |
-0.013 |
| 82 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.109 |
-0.123 |
| 83 |
266429_at |
AT2G07190
|
unknown |
-0.109 |
0.027 |
| 84 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.107 |
0.054 |
| 85 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.105 |
0.139 |
| 86 |
245095_at |
AT2G40860
|
[protein kinase family protein / protein phosphatase 2C ( PP2C) family protein] |
-0.104 |
-0.018 |
| 87 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
-0.104 |
0.012 |
| 88 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.104 |
-0.140 |
| 89 |
261427_at |
AT1G18670
|
IBS1, IMPAIRED IN BABA-INDUCED STERILITY 1 |
-0.102 |
-0.085 |
| 90 |
255184_at |
AT4G07730
|
[gypsy-like retrotransposon family (Athila), has a 6.8e-198 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.101 |
0.032 |
| 91 |
265076_at |
AT1G55490
|
CPN60B, chaperonin 60 beta, Cpn60beta1, chaperonin-60beta1, LEN1, LESION INITIATION 1 |
-0.099 |
0.002 |
| 92 |
261002_at |
AT1G26520
|
[Cobalamin biosynthesis CobW-like protein] |
-0.098 |
-0.055 |
| 93 |
257459_at |
AT1G24040
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.097 |
-0.030 |
| 94 |
257630_at |
AT3G26150
|
CYP71B16, cytochrome P450, family 71, subfamily B, polypeptide 16 |
-0.096 |
-0.041 |
| 95 |
256111_at |
AT1G16820
|
[vacuolar ATP synthase catalytic subunit-related / V-ATPase-related / vacuolar proton pump-related] |
-0.096 |
-0.035 |
| 96 |
266194_at |
AT2G39090
|
APC7, anaphase-promoting complex 7, AtAPC7 |
-0.095 |
-0.062 |
| 97 |
255234_at |
AT4G05500
|
[pseudogene] |
-0.095 |
0.047 |
| 98 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
-0.094 |
-0.189 |
| 99 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.092 |
-0.029 |
| 100 |
246875_at |
AT5G26130
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.087 |
0.043 |
| 101 |
245790_at |
AT1G32200
|
ACT1, ACYLTRANSFERASE 1, ATS1 |
-0.087 |
-0.042 |
| 102 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.085 |
-0.045 |
| 103 |
258853_at |
AT3G06440
|
[Galactosyltransferase family protein] |
-0.085 |
-0.124 |
| 104 |
264942_at |
AT1G67340
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.084 |
-0.059 |
| 105 |
246829_at |
AT5G26570
|
ATGWD3, OK1, PWD, PHOSPHOGLUCAN WATER DIKINASE |
-0.084 |
-0.045 |
| 106 |
265438_at |
AT2G20970
|
unknown |
-0.083 |
0.061 |
| 107 |
257057_at |
AT3G15310
|
unknown |
-0.083 |
0.059 |
| 108 |
263962_at |
AT2G36350
|
[Protein kinase superfamily protein] |
-0.082 |
-0.007 |
| 109 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
-0.081 |
0.091 |
| 110 |
257712_at |
AT3G27420
|
unknown |
-0.081 |
-0.133 |