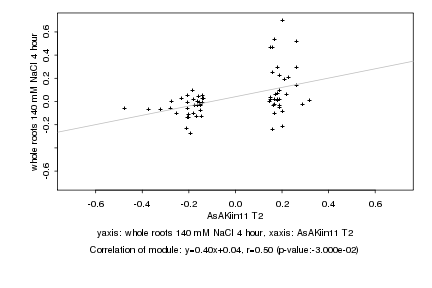
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
AsAKiin11 T2 |
Log2 signal ratio
whole roots 140 mM NaCl 4 hour |
| 1 |
263655_at |
AT1G04500
|
[CCT motif family protein] |
0.316 |
0.014 |
| 2 |
253178_at |
AT4G35170
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.284 |
-0.025 |
| 3 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.259 |
0.295 |
| 4 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.258 |
0.519 |
| 5 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.258 |
0.145 |
| 6 |
265711_at |
AT2G03360
|
[Glycosyltransferase family 61 protein] |
0.224 |
0.211 |
| 7 |
247458_at |
AT5G62180
|
AtCXE20, carboxyesterase 20, CXE20, carboxyesterase 20 |
0.215 |
0.064 |
| 8 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.209 |
0.195 |
| 9 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.202 |
0.707 |
| 10 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.200 |
-0.215 |
| 11 |
259338_at |
AT3G03800
|
ATSYP131, SYP131, syntaxin of plants 131 |
0.198 |
-0.085 |
| 12 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
0.189 |
0.025 |
| 13 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.189 |
-0.050 |
| 14 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.188 |
0.096 |
| 15 |
254469_at |
AT4G20470
|
unknown |
0.188 |
-0.028 |
| 16 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.186 |
0.232 |
| 17 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.180 |
0.298 |
| 18 |
250143_at |
AT5G14670
|
ARFA1B, ADP-ribosylation factor A1B, ATARFA1B, ADP-ribosylation factor A1B |
0.179 |
0.009 |
| 19 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.177 |
0.073 |
| 20 |
253699_at |
AT4G29800
|
PLA IVD, PLP8, PATATIN-like protein 8 |
0.177 |
0.023 |
| 21 |
248414_at |
AT5G51610
|
[Ribosomal protein L11 family protein] |
0.170 |
0.068 |
| 22 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.167 |
0.543 |
| 23 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
0.166 |
-0.095 |
| 24 |
266046_at |
AT2G07728
|
unknown |
0.165 |
-0.020 |
| 25 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
0.164 |
0.021 |
| 26 |
259634_at |
AT1G56380
|
[Mitochondrial transcription termination factor family protein] |
0.160 |
-0.032 |
| 27 |
261545_at |
AT1G63530
|
unknown |
0.159 |
-0.241 |
| 28 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.157 |
0.252 |
| 29 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.155 |
0.473 |
| 30 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
0.149 |
0.041 |
| 31 |
260495_at |
AT2G41810
|
unknown |
0.148 |
0.471 |
| 32 |
252451_at |
AT3G47100
|
unknown |
0.147 |
0.021 |
| 33 |
257031_at |
AT3G19280
|
ATFUT11, FUCT1, FUCTA, FUT11, fucosyltransferase 11 |
0.146 |
0.007 |
| 34 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
-0.477 |
-0.058 |
| 35 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.375 |
-0.065 |
| 36 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.322 |
-0.064 |
| 37 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.281 |
-0.057 |
| 38 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.277 |
0.002 |
| 39 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
-0.257 |
-0.096 |
| 40 |
258641_at |
AT3G08030
|
unknown |
-0.233 |
0.031 |
| 41 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.213 |
-0.232 |
| 42 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
-0.210 |
-0.052 |
| 43 |
248861_at |
AT5G46700
|
TET1, TETRASPANIN 1, TRN2, TORNADO 2 |
-0.207 |
0.056 |
| 44 |
266588_at |
AT2G14890
|
AGP9, arabinogalactan protein 9 |
-0.207 |
-0.002 |
| 45 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.206 |
-0.130 |
| 46 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.205 |
-0.109 |
| 47 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.204 |
-0.136 |
| 48 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.193 |
-0.269 |
| 49 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.186 |
0.100 |
| 50 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.182 |
-0.095 |
| 51 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.181 |
0.025 |
| 52 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.176 |
-0.028 |
| 53 |
263591_at |
AT2G01910
|
ATMAP65-6, MAP65-6 |
-0.168 |
-0.121 |
| 54 |
266889_at |
AT2G44640
|
unknown |
-0.166 |
0.005 |
| 55 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.165 |
-0.032 |
| 56 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.162 |
0.047 |
| 57 |
247770_at |
AT5G58930
|
unknown |
-0.155 |
-0.007 |
| 58 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.154 |
-0.069 |
| 59 |
265274_at |
AT2G28450
|
[zinc finger (CCCH-type) family protein] |
-0.151 |
-0.029 |
| 60 |
253558_at |
AT4G31120
|
ATPRMT5, PRMT5, PROTEIN ARGININE METHYLTRANSFERASE 5, SKB1, SHK1 binding protein 1 |
-0.151 |
-0.019 |
| 61 |
248273_at |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.146 |
-0.128 |
| 62 |
252990_at |
AT4G38440
|
IYO, MINIYO |
-0.144 |
0.032 |
| 63 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.144 |
0.055 |
| 64 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.142 |
-0.000 |
| 65 |
250908_at |
AT5G03680
|
PTL, PETAL LOSS |
-0.141 |
0.033 |