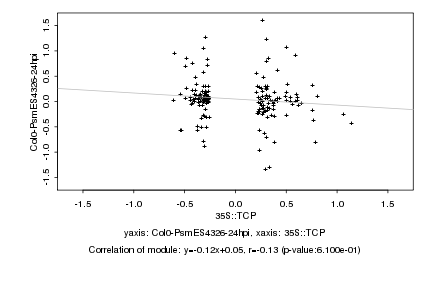
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::TCP |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
1.138 |
-0.426 |
| 2 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.056 |
-0.251 |
| 3 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.803 |
0.110 |
| 4 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.778 |
-0.797 |
| 5 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.761 |
-0.370 |
| 6 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
0.750 |
-0.168 |
| 7 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.749 |
0.334 |
| 8 |
245000_at |
ATCG00210
|
YCF6 |
0.646 |
-0.022 |
| 9 |
249010_at |
AT5G44580
|
unknown |
0.612 |
-0.068 |
| 10 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.606 |
0.086 |
| 11 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.603 |
0.035 |
| 12 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.599 |
0.154 |
| 13 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.584 |
0.928 |
| 14 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.583 |
0.017 |
| 15 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
0.557 |
-0.052 |
| 16 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.540 |
0.014 |
| 17 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.538 |
0.090 |
| 18 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.512 |
0.348 |
| 19 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.501 |
0.038 |
| 20 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
0.500 |
-0.258 |
| 21 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.500 |
1.069 |
| 22 |
246781_at |
AT5G27350
|
SFP1 |
0.498 |
0.193 |
| 23 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.491 |
0.118 |
| 24 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.429 |
0.071 |
| 25 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.408 |
0.069 |
| 26 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.407 |
0.623 |
| 27 |
249211_at |
AT5G42680
|
unknown |
0.405 |
0.019 |
| 28 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
0.391 |
0.031 |
| 29 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
0.383 |
-0.797 |
| 30 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.382 |
0.179 |
| 31 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
0.380 |
0.007 |
| 32 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.380 |
-0.291 |
| 33 |
258326_at |
AT3G22760
|
SOL1 |
0.373 |
-0.067 |
| 34 |
256926_at |
AT3G22540
|
unknown |
0.365 |
-0.144 |
| 35 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.365 |
-0.030 |
| 36 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
0.355 |
0.015 |
| 37 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.349 |
-0.265 |
| 38 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.341 |
0.036 |
| 39 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.335 |
-1.303 |
| 40 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.331 |
-0.129 |
| 41 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
0.326 |
0.116 |
| 42 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
0.323 |
-0.035 |
| 43 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
0.322 |
0.862 |
| 44 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
0.313 |
0.060 |
| 45 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.311 |
-0.308 |
| 46 |
257573_at |
AT2G33990
|
iqd9, IQ-domain 9 |
0.311 |
-0.176 |
| 47 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
0.309 |
0.299 |
| 48 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.309 |
-0.111 |
| 49 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.299 |
1.236 |
| 50 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.299 |
0.241 |
| 51 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.298 |
-0.707 |
| 52 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
0.298 |
0.259 |
| 53 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.298 |
0.125 |
| 54 |
260841_at |
AT1G29195
|
unknown |
0.296 |
0.795 |
| 55 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.292 |
-0.185 |
| 56 |
256096_at |
AT1G13650
|
unknown |
0.292 |
-1.342 |
| 57 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.292 |
0.303 |
| 58 |
264725_at |
AT1G22885
|
unknown |
0.287 |
0.078 |
| 59 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.281 |
-0.184 |
| 60 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.280 |
-0.628 |
| 61 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.272 |
-0.076 |
| 62 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.270 |
-0.212 |
| 63 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.269 |
-0.137 |
| 64 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.267 |
0.492 |
| 65 |
247107_at |
AT5G66040
|
STR16, sulfurtransferase protein 16 |
0.266 |
-0.119 |
| 66 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.265 |
1.620 |
| 67 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.262 |
0.011 |
| 68 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.261 |
0.217 |
| 69 |
245208_at |
AT5G12330
|
LRP1, LATERAL ROOT PRIMORDIUM 1 |
0.260 |
0.111 |
| 70 |
257697_at |
AT3G12700
|
NANA, NANA |
0.259 |
-0.239 |
| 71 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.256 |
0.022 |
| 72 |
246270_at |
AT4G36500
|
unknown |
0.249 |
0.259 |
| 73 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
0.247 |
-0.064 |
| 74 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
0.244 |
0.052 |
| 75 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
0.243 |
-0.021 |
| 76 |
261876_at |
AT1G50590
|
[RmlC-like cupins superfamily protein] |
0.232 |
-0.150 |
| 77 |
251563_at |
AT3G57880
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
0.231 |
0.282 |
| 78 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.230 |
-0.559 |
| 79 |
247457_at |
AT5G62170
|
TRM25, TON1 Recruiting Motif 25 |
0.230 |
-0.155 |
| 80 |
267354_at |
AT2G39880
|
AtMYB25, myb domain protein 25, MYB25, myb domain protein 25 |
0.228 |
-0.195 |
| 81 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.227 |
-0.964 |
| 82 |
260334_at |
AT1G70510
|
ATK1, ARABIDOPSIS THALIANA KN 1, KNAT2, KNOTTED-like from Arabidopsis thaliana 2 |
0.222 |
-0.018 |
| 83 |
252322_at |
AT3G48550
|
unknown |
0.222 |
-0.179 |
| 84 |
245007_at |
ATCG00350
|
PSAA |
0.221 |
-0.230 |
| 85 |
256335_at |
AT1G72110
|
[O-acyltransferase (WSD1-like) family protein] |
0.221 |
0.099 |
| 86 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
0.217 |
0.314 |
| 87 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
0.214 |
-0.232 |
| 88 |
247318_at |
AT5G63990
|
[Inositol monophosphatase family protein] |
0.207 |
0.559 |
| 89 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.206 |
0.187 |
| 90 |
258767_at |
AT3G10890
|
[Glycosyl hydrolase superfamily protein] |
-0.614 |
0.037 |
| 91 |
246145_at |
AT5G19880
|
[Peroxidase superfamily protein] |
-0.604 |
0.966 |
| 92 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.549 |
-0.557 |
| 93 |
259197_at |
AT3G03670
|
[Peroxidase superfamily protein] |
-0.549 |
0.150 |
| 94 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.531 |
-0.555 |
| 95 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
-0.497 |
0.707 |
| 96 |
256307_at |
AT1G30350
|
[Pectin lyase-like superfamily protein] |
-0.495 |
0.064 |
| 97 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.487 |
0.268 |
| 98 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.482 |
0.863 |
| 99 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.450 |
0.039 |
| 100 |
255717_at |
AT4G00350
|
[MATE efflux family protein] |
-0.448 |
0.080 |
| 101 |
267476_at |
AT2G02720
|
[Pectate lyase family protein] |
-0.434 |
-0.048 |
| 102 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.430 |
0.218 |
| 103 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
-0.428 |
0.761 |
| 104 |
264499_at |
AT1G30795
|
[Glycine-rich protein family] |
-0.423 |
-0.031 |
| 105 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.422 |
0.048 |
| 106 |
255276_at |
AT4G04930
|
DES-1-LIKE |
-0.412 |
0.153 |
| 107 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.406 |
0.009 |
| 108 |
261385_at |
AT1G05450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.402 |
0.232 |
| 109 |
264039_at |
AT2G03740
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.399 |
0.069 |
| 110 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.394 |
0.490 |
| 111 |
261528_at |
AT1G14420
|
AT59 |
-0.384 |
0.133 |
| 112 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.384 |
0.352 |
| 113 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.381 |
-0.563 |
| 114 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.378 |
-0.479 |
| 115 |
262817_at |
AT1G11770
|
[FAD-binding Berberine family protein] |
-0.370 |
0.054 |
| 116 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.368 |
0.047 |
| 117 |
261499_at |
AT1G28430
|
CYP705A24, cytochrome P450, family 705, subfamily A, polypeptide 24 |
-0.364 |
0.077 |
| 118 |
250251_at |
AT5G13670
|
UMAMIT15, Usually multiple acids move in and out Transporters 15 |
-0.364 |
-0.012 |
| 119 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
-0.358 |
0.101 |
| 120 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.357 |
-0.063 |
| 121 |
246437_at |
AT5G17540
|
[HXXXD-type acyl-transferase family protein] |
-0.354 |
0.122 |
| 122 |
248470_at |
AT5G50830
|
unknown |
-0.346 |
0.005 |
| 123 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.346 |
0.139 |
| 124 |
249760_at |
AT5G23960
|
ATTPS21, TPS21, terpene synthase 21 |
-0.337 |
0.086 |
| 125 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.336 |
-0.318 |
| 126 |
264987_at |
AT1G27030
|
unknown |
-0.335 |
-0.503 |
| 127 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.332 |
-0.062 |
| 128 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
-0.329 |
0.216 |
| 129 |
258468_at |
AT3G06070
|
unknown |
-0.323 |
-0.771 |
| 130 |
260553_at |
AT2G41800
|
unknown |
-0.321 |
-0.259 |
| 131 |
249541_at |
AT5G38130
|
[HXXXD-type acyl-transferase family protein] |
-0.321 |
0.306 |
| 132 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.319 |
0.044 |
| 133 |
251012_at |
AT5G02580
|
unknown |
-0.316 |
0.592 |
| 134 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.316 |
1.060 |
| 135 |
248229_at |
AT5G53810
|
[O-methyltransferase family protein] |
-0.315 |
-0.001 |
| 136 |
247804_at |
AT5G58170
|
GDPDL7, Glycerophosphodiester phosphodiesterase (GDPD) like 7, SVL5, SHV3-like 5 |
-0.315 |
0.094 |
| 137 |
251339_at |
AT3G60780
|
unknown |
-0.315 |
0.029 |
| 138 |
249323_at |
AT5G40940
|
FLA20, putative fasciclin-like arabinogalactan protein 20 |
-0.313 |
0.076 |
| 139 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
-0.312 |
-0.293 |
| 140 |
259464_at |
AT1G18990
|
unknown |
-0.311 |
0.004 |
| 141 |
261943_at |
AT1G80660
|
AHA9, H(+)-ATPase 9, HA9, H(+)-ATPase 9 |
-0.310 |
0.134 |
| 142 |
246592_at |
AT5G14890
|
[NHL domain-containing protein] |
-0.309 |
0.166 |
| 143 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
-0.309 |
0.090 |
| 144 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.307 |
-0.877 |
| 145 |
265368_at |
AT2G13350
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.307 |
0.045 |
| 146 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.304 |
0.311 |
| 147 |
247424_at |
AT5G62850
|
AtSWEET5, AtVEX1, VEGETATIVE CELL EXPRESSED1, SWEET5 |
-0.303 |
0.201 |
| 148 |
245036_at |
AT2G26410
|
Iqd4, IQ-domain 4 |
-0.302 |
0.061 |
| 149 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.301 |
1.271 |
| 150 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.298 |
-0.138 |
| 151 |
252140_at |
AT3G51070
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.296 |
0.038 |
| 152 |
265127_at |
AT1G55560
|
sks14, SKU5 similar 14 |
-0.296 |
0.051 |
| 153 |
263028_at |
AT1G24030
|
[Protein kinase superfamily protein] |
-0.294 |
0.193 |
| 154 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.291 |
-0.497 |
| 155 |
257399_at |
AT1G23690
|
unknown |
-0.290 |
-0.029 |
| 156 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.287 |
-0.297 |
| 157 |
257256_at |
AT3G21970
|
unknown |
-0.287 |
0.080 |
| 158 |
262463_at |
AT1G50310
|
ATSTP9, SUGAR TRANSPORTER 9, STP9, sugar transporter 9 |
-0.286 |
0.155 |
| 159 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.283 |
0.715 |
| 160 |
254123_at |
AT4G24640
|
APPB1 |
-0.283 |
0.011 |
| 161 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.283 |
0.844 |
| 162 |
251595_at |
AT3G57620
|
[glyoxal oxidase-related protein] |
-0.281 |
0.050 |
| 163 |
251250_at |
AT3G62180
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.277 |
0.070 |
| 164 |
253151_at |
AT4G35670
|
[Pectin lyase-like superfamily protein] |
-0.276 |
0.027 |
| 165 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
-0.275 |
0.104 |
| 166 |
264993_at |
AT1G67290
|
GLOX1, glyoxal oxidase 1 |
-0.274 |
0.076 |
| 167 |
258671_at |
AT3G08560
|
VHA-E2, vacuolar H+-ATPase subunit E isoform 2 |
-0.273 |
0.070 |
| 168 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.272 |
-0.011 |
| 169 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.271 |
0.214 |
| 170 |
260888_at |
AT1G29140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.270 |
0.065 |
| 171 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
-0.270 |
0.004 |
| 172 |
267449_at |
AT2G33690
|
[Late embryogenesis abundant protein, group 6] |
-0.269 |
0.111 |
| 173 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.268 |
0.314 |
| 174 |
257439_at |
AT2G17000
|
[Mechanosensitive ion channel family protein] |
-0.268 |
0.044 |
| 175 |
261244_at |
AT1G20150
|
[Subtilisin-like serine endopeptidase family protein] |
-0.267 |
0.093 |
| 176 |
249852_at |
AT5G23270
|
ATSTP11, SUGAR TRANSPORTER 11, STP11, sugar transporter 11 |
-0.266 |
0.072 |
| 177 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.264 |
-0.301 |