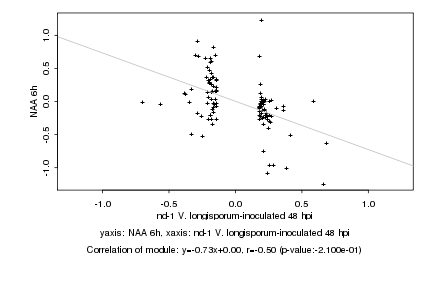
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
nd-1 V. longisporum-inoculated 48 hpi |
Log2 signal ratio
NAA 6h |
| 1 |
266222_at |
AT2G28780
|
unknown |
0.682 |
-0.634 |
| 2 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
0.658 |
-1.239 |
| 3 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.584 |
0.011 |
| 4 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.413 |
-0.501 |
| 5 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.380 |
-1.001 |
| 6 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.360 |
-0.135 |
| 7 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.358 |
-0.069 |
| 8 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.306 |
-0.091 |
| 9 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.284 |
-0.960 |
| 10 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.271 |
0.028 |
| 11 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.264 |
-0.224 |
| 12 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
0.257 |
-0.307 |
| 13 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.255 |
-0.289 |
| 14 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.254 |
-0.965 |
| 15 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.253 |
-0.204 |
| 16 |
248706_at |
AT5G48530
|
unknown |
0.251 |
0.005 |
| 17 |
250095_at |
AT5G17230
|
PSY, PHYTOENE SYNTHASE |
0.246 |
-0.405 |
| 18 |
250127_at |
AT5G16380
|
unknown |
0.241 |
-0.259 |
| 19 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.238 |
-1.077 |
| 20 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
0.237 |
-0.226 |
| 21 |
246450_at |
AT5G16820
|
ATHSF3, ARABIDOPSIS HEAT SHOCK FACTOR 3, ATHSFA1B, ARABIDOPSIS THALIANA CLASS A HEAT SHOCK FACTOR 1B, HSF3, heat shock factor 3, HSFA1B, CLASS A HEAT SHOCK FACTOR 1B |
0.227 |
-0.176 |
| 22 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.223 |
-0.227 |
| 23 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
0.222 |
-0.210 |
| 24 |
250117_at |
AT5G16440
|
IPP1, isopentenyl diphosphate isomerase 1 |
0.222 |
0.044 |
| 25 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.217 |
0.005 |
| 26 |
246486_at |
AT5G15910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.215 |
-0.109 |
| 27 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
0.212 |
-0.134 |
| 28 |
250113_at |
AT5G16320
|
FRL1, FRIGIDA like 1 |
0.211 |
-0.030 |
| 29 |
253724_at |
AT4G29285
|
LCR24, low-molecular-weight cysteine-rich 24 |
0.208 |
-0.008 |
| 30 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
0.208 |
-0.036 |
| 31 |
250164_at |
AT5G15280
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.207 |
0.026 |
| 32 |
246482_at |
AT5G15930
|
PAM1, plant adhesion molecule 1 |
0.204 |
-0.241 |
| 33 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.204 |
-0.750 |
| 34 |
256796_at |
AT3G22210
|
unknown |
0.204 |
-0.341 |
| 35 |
254747_at |
AT4G13020
|
MHK |
0.199 |
-0.248 |
| 36 |
250122_at |
AT5G16520
|
unknown |
0.199 |
-0.126 |
| 37 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
0.195 |
0.073 |
| 38 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
0.195 |
-0.069 |
| 39 |
244901_at |
ATMG00640
|
ORF25 |
0.194 |
0.003 |
| 40 |
250074_at |
AT5G17310
|
AtUGP2, UDP-GLUCOSE PYROPHOSPHORYLASE 2, UGP2, UDP-glucose pyrophosphorylase 2 |
0.192 |
-0.172 |
| 41 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.191 |
1.231 |
| 42 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
0.190 |
-0.017 |
| 43 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.189 |
0.040 |
| 44 |
246554_at |
AT5G15450
|
APG6, ALBINO AND PALE GREEN 6, AtCLPB3, CLPB-P, CASEIN LYTIC PROTEINASE B-P, CLPB3, casein lytic proteinase B3 |
0.189 |
0.003 |
| 45 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
0.186 |
-0.225 |
| 46 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.185 |
0.262 |
| 47 |
246561_at |
AT5G15570
|
[Bromodomain transcription factor] |
0.184 |
0.136 |
| 48 |
246553_at |
AT5G15440
|
EDL1, EID1-like 1 |
0.184 |
-0.042 |
| 49 |
246513_at |
AT5G15680
|
[ARM repeat superfamily protein] |
0.183 |
-0.087 |
| 50 |
246422_at |
AT5G17060
|
ARFB1B, ADP-ribosylation factor B1B, ATARFB1B, ADP-ribosylation factor B1B |
0.182 |
-0.022 |
| 51 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
0.180 |
-0.203 |
| 52 |
246449_at |
AT5G16810
|
[Protein kinase superfamily protein] |
0.180 |
-0.105 |
| 53 |
246453_at |
AT5G16830
|
ATPEP12, ATSYP21, PEP12, PEP12P, SYP21, syntaxin of plants 21 |
0.178 |
-0.140 |
| 54 |
250563_at |
AT5G08050
|
unknown |
0.177 |
-0.266 |
| 55 |
246515_at |
AT5G15710
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.175 |
-0.080 |
| 56 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
0.174 |
-0.089 |
| 57 |
246539_at |
AT5G15460
|
MUB2, membrane-anchored ubiquitin-fold protein 2 |
0.174 |
-0.074 |
| 58 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.174 |
0.685 |
| 59 |
251755_at |
AT3G55790
|
unknown |
-0.704 |
-0.008 |
| 60 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.570 |
-0.033 |
| 61 |
266469_at |
AT2G31180
|
ATMYB14, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 14, MYB14, myb domain protein 14, MYB14AT |
-0.388 |
0.130 |
| 62 |
248556_at |
AT5G50350
|
unknown |
-0.383 |
0.117 |
| 63 |
259028_at |
AT3G09290
|
TAC1, telomerase activator1 |
-0.348 |
-0.005 |
| 64 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.335 |
0.186 |
| 65 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.331 |
-0.486 |
| 66 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.303 |
0.709 |
| 67 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.292 |
0.918 |
| 68 |
262719_at |
AT1G43590
|
unknown |
-0.286 |
-0.171 |
| 69 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.281 |
0.685 |
| 70 |
266118_at |
AT2G02130
|
LCR68, low-molecular-weight cysteine-rich 68, PDF2.3 |
-0.256 |
-0.223 |
| 71 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.254 |
-0.525 |
| 72 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.227 |
0.664 |
| 73 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
-0.220 |
0.368 |
| 74 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.216 |
0.150 |
| 75 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.215 |
-0.020 |
| 76 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.212 |
0.515 |
| 77 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.209 |
-0.257 |
| 78 |
249911_at |
AT5G22740
|
ATCSLA02, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE A02, ATCSLA2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE A2, CSLA02, cellulose synthase-like A02, CSLA2, CELLULOSE SYNTHASE-LIKE A 2 |
-0.204 |
0.328 |
| 79 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.203 |
0.063 |
| 80 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
-0.200 |
0.471 |
| 81 |
257465_at |
AT1G13040
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.200 |
0.285 |
| 82 |
266865_at |
AT2G29980
|
AtFAD3, FAD3, fatty acid desaturase 3 |
-0.200 |
0.302 |
| 83 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
-0.195 |
0.661 |
| 84 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.194 |
-0.198 |
| 85 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.194 |
-0.202 |
| 86 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
-0.194 |
0.601 |
| 87 |
254112_at |
AT4G24970
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
-0.191 |
0.335 |
| 88 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.186 |
0.266 |
| 89 |
249377_at |
AT5G40690
|
unknown |
-0.185 |
-0.266 |
| 90 |
253492_at |
AT4G31810
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.183 |
0.435 |
| 91 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.181 |
0.607 |
| 92 |
255543_at |
AT4G01870
|
[tolB protein-related] |
-0.181 |
0.044 |
| 93 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
-0.181 |
0.140 |
| 94 |
253131_at |
AT4G35550
|
ATWOX13, HB-4, WOX13, WUSCHEL related homeobox 13 |
-0.179 |
0.369 |
| 95 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
-0.179 |
0.157 |
| 96 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
-0.178 |
-0.111 |
| 97 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.177 |
-0.347 |
| 98 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
-0.172 |
0.227 |
| 99 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
-0.171 |
-0.019 |
| 100 |
248132_at |
AT5G54840
|
ATSGP1, SGP1 |
-0.170 |
-0.081 |
| 101 |
262943_at |
AT1G79470
|
[Aldolase-type TIM barrel family protein] |
-0.170 |
0.366 |
| 102 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.170 |
0.827 |
| 103 |
254340_at |
AT4G22120
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.167 |
-0.161 |
| 104 |
251401_at |
AT3G60270
|
[Cupredoxin superfamily protein] |
-0.162 |
-0.044 |
| 105 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.155 |
0.032 |
| 106 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
-0.154 |
0.702 |
| 107 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.153 |
0.157 |
| 108 |
255252_at |
AT4G04990
|
unknown |
-0.150 |
-0.259 |
| 109 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.149 |
-0.028 |
| 110 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
-0.148 |
0.346 |
| 111 |
265082_at |
AT1G03830
|
[guanylate-binding family protein] |
-0.148 |
0.223 |
| 112 |
261463_at |
AT1G07740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.146 |
0.174 |
| 113 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
-0.145 |
0.159 |
| 114 |
252001_at |
AT3G52750
|
FTSZ2-2 |
-0.144 |
-0.075 |
| 115 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
-0.143 |
0.323 |