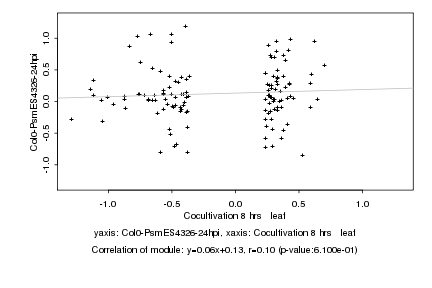
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Cocultivation 8 hrs leaf |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.699 |
0.585 |
| 2 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.643 |
0.034 |
| 3 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.616 |
0.953 |
| 4 |
260744_at |
AT1G15010
|
unknown |
0.593 |
0.435 |
| 5 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.585 |
-0.086 |
| 6 |
257670_at |
AT3G20340
|
unknown |
0.584 |
0.300 |
| 7 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.521 |
-0.845 |
| 8 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.450 |
0.057 |
| 9 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.432 |
0.984 |
| 10 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.427 |
0.085 |
| 11 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.422 |
0.293 |
| 12 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.421 |
0.279 |
| 13 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.416 |
0.813 |
| 14 |
264238_at |
AT1G54740
|
unknown |
0.406 |
-0.349 |
| 15 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.402 |
0.063 |
| 16 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.391 |
0.227 |
| 17 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.389 |
0.658 |
| 18 |
255733_at |
AT1G25400
|
unknown |
0.378 |
0.742 |
| 19 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.373 |
-0.449 |
| 20 |
266798_at |
AT2G22850
|
AtbZIP6, basic leucine-zipper 6, bZIP6, basic leucine-zipper 6 |
0.372 |
0.399 |
| 21 |
250223_at |
AT5G14070
|
ROXY2 |
0.361 |
0.030 |
| 22 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.358 |
-0.085 |
| 23 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.358 |
-0.582 |
| 24 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.354 |
0.171 |
| 25 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.341 |
0.009 |
| 26 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.331 |
0.390 |
| 27 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.329 |
0.370 |
| 28 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.328 |
0.278 |
| 29 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.327 |
-0.135 |
| 30 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.326 |
0.495 |
| 31 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.326 |
-0.087 |
| 32 |
263118_at |
AT1G03090
|
MCCA |
0.322 |
0.322 |
| 33 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.319 |
0.960 |
| 34 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.316 |
0.804 |
| 35 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.308 |
0.192 |
| 36 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.303 |
0.697 |
| 37 |
259103_at |
AT3G11690
|
unknown |
0.302 |
-0.125 |
| 38 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.302 |
0.034 |
| 39 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.298 |
0.397 |
| 40 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
0.295 |
0.016 |
| 41 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.294 |
0.043 |
| 42 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.288 |
-0.705 |
| 43 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.287 |
-0.433 |
| 44 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.280 |
-0.274 |
| 45 |
262026_at |
AT1G35670
|
ATCDPK2, calcium-dependent protein kinase 2, ATCPK11, CDPK2, calcium-dependent protein kinase 2, CPK11 |
0.280 |
0.069 |
| 46 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.279 |
0.697 |
| 47 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.278 |
0.261 |
| 48 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.276 |
0.216 |
| 49 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
0.273 |
0.075 |
| 50 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.273 |
0.736 |
| 51 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
0.272 |
0.075 |
| 52 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.272 |
-0.150 |
| 53 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.268 |
0.097 |
| 54 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
0.262 |
-0.023 |
| 55 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.261 |
0.258 |
| 56 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.260 |
0.110 |
| 57 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.260 |
0.888 |
| 58 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.256 |
-0.189 |
| 59 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.253 |
0.178 |
| 60 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.249 |
0.272 |
| 61 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.239 |
-0.387 |
| 62 |
256799_at |
AT3G18560
|
unknown |
0.236 |
0.445 |
| 63 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.236 |
0.038 |
| 64 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.234 |
-0.721 |
| 65 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.234 |
0.456 |
| 66 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.233 |
-0.138 |
| 67 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.233 |
-0.275 |
| 68 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.232 |
-0.574 |
| 69 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-1.296 |
-0.284 |
| 70 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.141 |
0.199 |
| 71 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-1.120 |
0.334 |
| 72 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-1.119 |
0.101 |
| 73 |
249598_at |
AT5G37970
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.062 |
0.017 |
| 74 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-1.052 |
-0.308 |
| 75 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-1.008 |
0.071 |
| 76 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.965 |
-0.036 |
| 77 |
252063_at |
AT3G51590
|
LTP12, lipid transfer protein 12 |
-0.881 |
0.095 |
| 78 |
260038_at |
AT1G68875
|
unknown |
-0.877 |
0.041 |
| 79 |
261684_at |
AT1G47400
|
unknown |
-0.870 |
-0.097 |
| 80 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.834 |
0.883 |
| 81 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.775 |
1.031 |
| 82 |
260001_at |
AT1G67990
|
ATTSM1, TAPETUM-SPECIFIC METHYLTRANSFERASE 1, TSM1 |
-0.768 |
0.113 |
| 83 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.757 |
0.125 |
| 84 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.750 |
0.627 |
| 85 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.722 |
0.096 |
| 86 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.692 |
0.035 |
| 87 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.691 |
0.027 |
| 88 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.674 |
1.071 |
| 89 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.659 |
0.536 |
| 90 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.654 |
0.017 |
| 91 |
254574_at |
AT4G19430
|
unknown |
-0.641 |
0.106 |
| 92 |
262697_at |
AT1G75940
|
ATA27, BGLU20, BETA GLUCOSIDASE 20 |
-0.637 |
0.025 |
| 93 |
264635_at |
AT1G65500
|
unknown |
-0.616 |
-0.182 |
| 94 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.597 |
0.487 |
| 95 |
249614_at |
AT5G37300
|
WSD1 |
-0.593 |
-0.791 |
| 96 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.572 |
-0.120 |
| 97 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
-0.572 |
0.122 |
| 98 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.568 |
0.138 |
| 99 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.567 |
0.176 |
| 100 |
257421_at |
AT1G12030
|
unknown |
-0.555 |
0.045 |
| 101 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.540 |
-0.040 |
| 102 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.526 |
-0.436 |
| 103 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.522 |
0.405 |
| 104 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.519 |
0.225 |
| 105 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.515 |
-0.514 |
| 106 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.511 |
1.064 |
| 107 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
-0.510 |
0.942 |
| 108 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.504 |
0.116 |
| 109 |
250828_at |
AT5G05250
|
unknown |
-0.502 |
-0.064 |
| 110 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.490 |
-0.085 |
| 111 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.484 |
-0.707 |
| 112 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.479 |
0.065 |
| 113 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.478 |
0.323 |
| 114 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.478 |
-0.049 |
| 115 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.471 |
-0.674 |
| 116 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.455 |
0.308 |
| 117 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.439 |
-0.091 |
| 118 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.436 |
-0.142 |
| 119 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.429 |
-0.114 |
| 120 |
263829_at |
AT2G40435
|
unknown |
-0.427 |
0.116 |
| 121 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.426 |
0.381 |
| 122 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.415 |
0.123 |
| 123 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.413 |
-0.059 |
| 124 |
259372_at |
AT1G69120
|
AGL7, AGAMOUS-like 7, AP1, APETALA1, AtAP1 |
-0.408 |
-0.001 |
| 125 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.394 |
1.192 |
| 126 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.393 |
0.077 |
| 127 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
-0.393 |
0.354 |
| 128 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.387 |
0.148 |
| 129 |
255912_at |
AT1G66960
|
LUP5 |
-0.387 |
-0.159 |
| 130 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
-0.387 |
0.073 |
| 131 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.385 |
-0.803 |
| 132 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
-0.385 |
-0.149 |
| 133 |
261221_at |
AT1G19960
|
unknown |
-0.380 |
-0.409 |
| 134 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.374 |
0.081 |
| 135 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.366 |
0.406 |