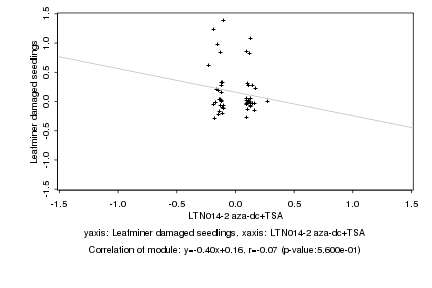
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
LTN014-2 aza-dc+TSA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.270 |
0.017 |
| 2 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.168 |
0.236 |
| 3 |
256811_at |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
0.159 |
-0.023 |
| 4 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.156 |
-0.138 |
| 5 |
246589_at |
AT5G14860
|
[UDP-Glycosyltransferase superfamily protein] |
0.145 |
-0.026 |
| 6 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.143 |
0.284 |
| 7 |
251254_at |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
0.127 |
-0.028 |
| 8 |
245669_at |
AT1G28300
|
LEC2, LEAFY COTYLEDON 2 |
0.127 |
-0.075 |
| 9 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.126 |
1.094 |
| 10 |
265688_at |
AT2G24300
|
[Calmodulin-binding protein] |
0.124 |
-0.066 |
| 11 |
246727_at |
AT5G28010
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.124 |
0.060 |
| 12 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.116 |
0.826 |
| 13 |
265316_at |
AT2G20400
|
[myb-like HTH transcriptional regulator family protein] |
0.113 |
0.002 |
| 14 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.113 |
-0.008 |
| 15 |
244931_at |
ATMG00630
|
ORF110B |
0.108 |
0.020 |
| 16 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.108 |
0.288 |
| 17 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
0.103 |
-0.008 |
| 18 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.102 |
0.313 |
| 19 |
266305_at |
AT2G27120
|
POL2B, TIL2, TILTED 2 |
0.101 |
-0.002 |
| 20 |
263370_at |
AT2G20500
|
unknown |
0.099 |
-0.032 |
| 21 |
255402_at |
AT4G03205
|
hemf2 |
0.096 |
-0.131 |
| 22 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
0.094 |
-0.271 |
| 23 |
252650_at |
AT3G44690
|
unknown |
0.094 |
0.020 |
| 24 |
258718_at |
AT3G09760
|
[RING/U-box superfamily protein] |
0.094 |
-0.042 |
| 25 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.092 |
0.052 |
| 26 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.090 |
0.868 |
| 27 |
257346_at |
AT3G30846
|
[gypsy-like retrotransposon family, has a 1.0e-303 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.087 |
-0.023 |
| 28 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
-0.236 |
0.626 |
| 29 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.193 |
1.239 |
| 30 |
267480_at |
AT2G02830
|
[copia-like retrotransposon family, has a 1.3e-37 P-value blast match to GB:CAA72990 open reading frame 2 (Ty1_Copia-element) (Brassica oleracea)] |
-0.192 |
-0.037 |
| 31 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.182 |
-0.289 |
| 32 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.173 |
-0.016 |
| 33 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
-0.165 |
0.216 |
| 34 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
-0.155 |
0.989 |
| 35 |
252108_at |
AT3G51530
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.149 |
-0.208 |
| 36 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.146 |
0.189 |
| 37 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.144 |
0.048 |
| 38 |
258548_at |
AT3G06910
|
AtULP1a, ELS1, ESD4-LIKE SUMO PROTEASE 1, ULP1A, UB-like protease 1A |
-0.138 |
-0.160 |
| 39 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.131 |
0.843 |
| 40 |
264870_at |
AT1G24210
|
[Paired amphipathic helix (PAH2) superfamily protein] |
-0.131 |
0.050 |
| 41 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
-0.131 |
-0.055 |
| 42 |
257672_at |
AT3G20300
|
unknown |
-0.126 |
0.031 |
| 43 |
263439_at |
AT2G28650
|
ATEXO70H8, exocyst subunit exo70 family protein H8, EXO70H8, exocyst subunit exo70 family protein H8 |
-0.126 |
0.163 |
| 44 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
-0.123 |
0.289 |
| 45 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.121 |
0.009 |
| 46 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-0.119 |
0.328 |
| 47 |
255449_at |
AT4G02820
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.116 |
-0.188 |
| 48 |
247515_at |
AT5G61740
|
ABCA10, ATP-binding cassette A10, ATATH14, ARABIDOPSIS THALIANA ABC2 HOMOLOG 14, ATH14, ABC2 homolog 14 |
-0.114 |
-0.094 |
| 49 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
-0.111 |
0.328 |
| 50 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.109 |
-0.058 |
| 51 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.107 |
1.403 |
| 52 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.107 |
-0.117 |
| 53 |
246433_at |
AT5G17510
|
unknown |
-0.103 |
-0.113 |